Team:Imperial College London/Modelling
From 2010.igem.org
(Difference between revisions)
m (Change Display Protein Model into Sruface protein and receptor model) |
|||
| Line 8: | Line 8: | ||
<ol> | <ol> | ||
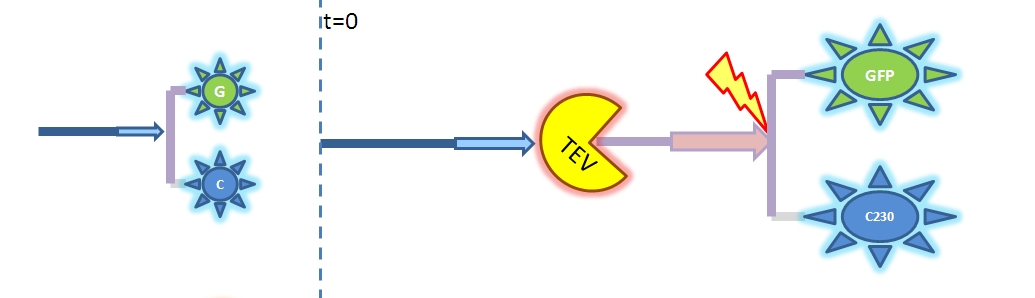
<li><b>Output Amplification Model</b><br/>We came up with an idea of using amplification of colour output to make it show within minutes after stimulus. The question that arose concerned whether amplification will actually perform better than simple production in the cellular environment. Furthermore, we had trouble deciding whether we should design the amplification module to be consisting of 1,2 or even more amplification steps. It appeared that the problem was recognised to be difficult enough to employ modelling.</li> | <li><b>Output Amplification Model</b><br/>We came up with an idea of using amplification of colour output to make it show within minutes after stimulus. The question that arose concerned whether amplification will actually perform better than simple production in the cellular environment. Furthermore, we had trouble deciding whether we should design the amplification module to be consisting of 1,2 or even more amplification steps. It appeared that the problem was recognised to be difficult enough to employ modelling.</li> | ||
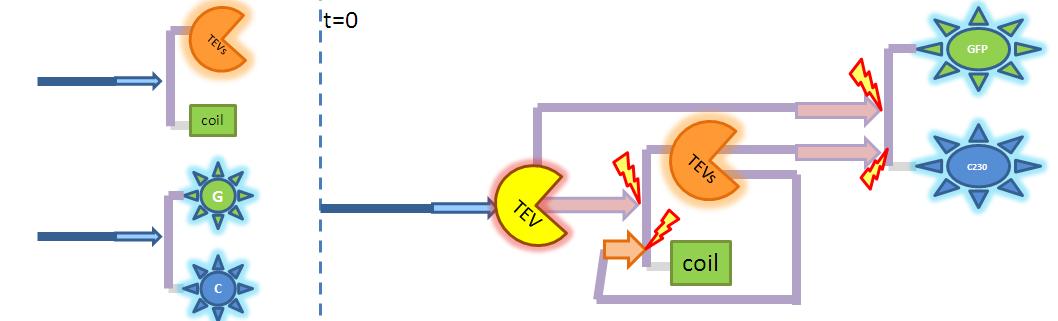
| - | <li><b> | + | <li><b>Protein Display Model</b><br/>We came up with novel idea of detecting organisms that we do not have a specific receptor for. In the particular example that we have been considering, Schistosoma's protease was meant to cleave designed by us protein displayed on bacteria's cell wall. The cleaved peptide was supposed to be recognised by the receptor which would act to activate the colour expression. That solution raised questions about the risk of false positive or whether in there are any chances for ComD receptors to get activated in the diluted environment. Modelling was recognised as suitable to answer those questions.</li> |
</ol> | </ol> | ||
|} | |} | ||
| Line 35: | Line 35: | ||
| - | <b style="font-size:12px"> | + | <b style="font-size:12px">Protein Display Model</b><br/> |
<ol> | <ol> | ||
<li>Initial TEV protease concentrations we determined for the optimal activation of the receptor within 1.5 minute after elastases would have come into contact with our cell.</li> | <li>Initial TEV protease concentrations we determined for the optimal activation of the receptor within 1.5 minute after elastases would have come into contact with our cell.</li> | ||
| Line 88: | Line 88: | ||
| - | <b style="font-size:12px"> | + | <b style="font-size:12px">Protein Display Model</b><br/> |
Goals: | Goals: | ||
<ol><p>The aim of this model is to determine the concentration of Schistosoma elastase or TEV protease that should be added to bacteria to trigger the response.</p> | <ol><p>The aim of this model is to determine the concentration of Schistosoma elastase or TEV protease that should be added to bacteria to trigger the response.</p> | ||
| Line 95: | Line 95: | ||
Elements of the system: | Elements of the system: | ||
| - | <ol><li> | + | <ol><li>Display protein that consists of cell wall binding domain, linker, AIP (Auto Inducing Peptide)</li> |
<li>Schistosoma elastase (enzyme released by the parasite) cleaves AIP from cell wall binding domain at the linker site. In laboratory we used TEV protease as we could not get handle of Schistosoma elastase.</li> | <li>Schistosoma elastase (enzyme released by the parasite) cleaves AIP from cell wall binding domain at the linker site. In laboratory we used TEV protease as we could not get handle of Schistosoma elastase.</li> | ||
<li>ComD receptor is being activated by high enough AIP concentration.</li> | <li>ComD receptor is being activated by high enough AIP concentration.</li> | ||
Revision as of 20:17, 12 October 2010
| Introduction to modelling |
In the process of designing our construct two major questions arose which could be answered by computer modelling:
|
| Results & Conclusions | ||||
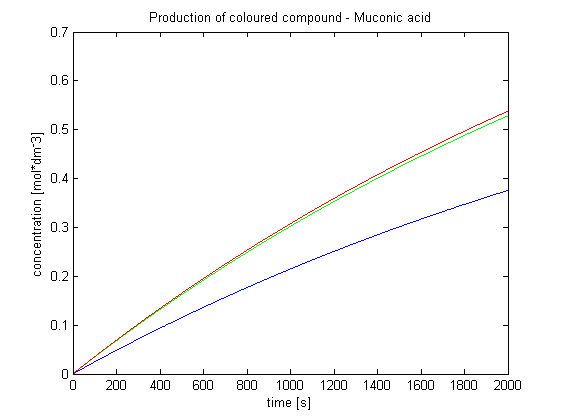
Output Amplification Model
|
| Quick overview of models | ||||||
| Output Amplification Model Goals: This model was mainly developed in order to determine whether simple production is better than 1- or 2-step amplification. Further goals, contained estimation of the speed of modelled response.Elements of the system:
Major assumptions:
The aim of this model is to determine the concentration of Schistosoma elastase or TEV protease that should be added to bacteria to trigger the response. It is also attempted to model how long it takes for the protease or elastase to cleave enough peptides. Elements of the system:
Major assumptions:
|
 "
"