Team:Lethbridge/Notebook/Lab Work/June
From 2010.igem.org
Adam.smith4 (Talk | contribs) (→June 2010) |
JustinVigar (Talk | contribs) (→June 3/2010 - Evening) |
||
| Line 210: | Line 210: | ||
===June 3/2010 - Evening=== | ===June 3/2010 - Evening=== | ||
| - | <b>Objective:</b> Repeat restriction of pSB1T3 and ligate with pLacI and sRBS-Lum-dT. Previous ligations all used up on gel. | + | <b>Objective:</b> Repeat restriction of pSB1T3 and ligate with pLacI and sRBS-Lum-dT. Previous ligations all used up on gel.<br> |
<b>Method:</b><br> | <b>Method:</b><br> | ||
Ligation:<br> | Ligation:<br> | ||
Revision as of 17:04, 11 June 2010
Contents |
June 2010
June 1/2010
JV quantified the amount of DNA in gels run to date using ImageJ software. Results to be posted in working plasmids box.
Objective: Transform plasmids into DH5α
Method: Follow competent cell transformation protocol to transform the following:
From our ligations:
- pLacI-sRBS-Lumazine-dT
- pLacI-sRBS-Lumazine-dT
- mms6 (A6)
- mms6 (B6)
- xylE (C4)
- xylE (B4)
From the 2010 Parts Distribution:
- ECFP (Bba_E0020)
- EYFP (Bba_E0030)
- BglII Endonuclease (Bba_K112106)
June 2/2010
(In Lab: JV)
Objective: Isolate plasmid DNA of RBS-xylE (BBa_J33204) from DH5α cells and confirm results.
Method: "Mini-prep" the plasmid DNA using boiling lysis miniprep. Then restrict the DNA once and run on a 1% agarose gel (TAE).
Restriction Reaction
| Ingredient | Volume(µL) |
| MilliQ H20 Water | 15.75 |
| Orange Buffer (10x) | 2 |
| pDNA (rbs-xylE) | 2 |
| EcoRI | 0.25 |
Unrestricted Control
| Ingredient | Volume(µL) |
| MilliQ H20 Water | 16 |
| Orange Buffer (10x) | 2 |
| pDNA (rbs-xylE) | 2 |
DNA was restricted for 80 minutes at 37oC.
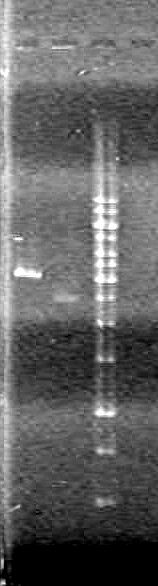
Analyzed results on a 1% agarose gel. Load order as follows:
| Lane | Sample | Volume Sample (µL) | Volume Loading Dye (µL) |
| 1 | Restricted RBS-xylE | 10 | 2 |
| 1 | Unestricted RBS-xylE† | 1 | 2 |
| 1 | 1kb Ladder†† | 2 | 2 |
† Added 9µL MilliQ H2O
†† Added 8µL MilliQ H2O
Ran gel at 100V from 2 hours.
Results:
Conclusions: Plasmid DNA prep and restriction was successful.
Objective: Ligate rbs-xylE (Bba_J33204) to our double terminator, and insert it into the pSB1T3 plasmid backbone.
Method:
- Restrictions
- Restrict rbs-xylE wit EcoRI and SpeI (Red Buffer)
- Restrict the double terminator with XbaI and PstI (Tango Buffer)
- Restrict pSB1T3 with EcoRI and PstI (Red Buffer)
Component Volume (µL) MilliQ H2O 15.5 Buffer 2 pDNA 2 Enzyme 0.25 + 0.25 Set up control reaction as follows:
- MilliQ H2O - 16µL
- Buffer - 2µL
- pDNA - 2µL
Incubated reactions for 65 minutes at 37oC
Killed enzymes by incubating reactions for 10 minutes at 65oC
- Ligation
Reaction set up as follows:- T4 DNA ligase - 0.25µL
- rbs-xylE - 5µL
- dT - 3µL
- pSB1T3 - 8µL
- 10x Ligation Buffer - 2µL
- MilliQ H2O - 1.75µL
Killed enzymes by incubating reactions for 10 minutes at 80oC</ul>June 2/2010 - Evening
Objective: Set up new ligations of pLacI and sRBS-Lum-dT according to Tom Knight's protocol. Previous ligation had very little DNA.
Relevant Information:
- Want a final mass of 25ng of each pDNA in the ligation mix.
- Final concentration of pDNA in restriction digest should be 25-50ng/µL.
- Tom Knight's restriction reaction is 50µL, therefore there should be 1000ng pDNA in each restriction digest.
- Identified the following plasmids in our working plasmids box:
Common Name Location Concentration (ng/µL) Volume/rxn (µL) pLacI Maxiprep A9 990 ~1 pLacI (B1) A6 440 ~2 sRBS-Lum-dT (2) A1 965 ~1 sRBS-Lum-dT (1) A2 1145 ~1 sRBS-Lum-dT Maxiprep B8 4780 ~.2 sRBS-Lum-dT B7 4375 ~.25 sRBS-Lum-dT (1) G2 335 ~3 sRBS-Lum-dT (2) G3 965 ~2 - Make a 1:10 dilution of sRBS-Lum-dt maxiprep (D8) and sRBS-Lum-dT (B7). 0.5µL pDNA in 4.5µL water.
- Cut pLacI with EcoRI and SpeI
- Cut sRBS-Lum-dT with XbaI and PstI
- Cut pSB1T3 with EcoRI and PstI
- Will have total of 12 ligation reactions, want 12x2µL of pSB1T3 to add to each, therefore want 25µL of pSB1T3.
Method:
Restriction
Name [pDNA] (ng/µL) Volume
pDNA (µL)Volume
Water (µL)Volume
Buffer (µL)Enzymes Total Volume sRBS-Lum-dT (A1) 965 1 43.5 5 0.25µL XbaI
0.25µL PstI50 sRBS-Lum-dT (A2) 1145 1 43.5 5 0.25µL XbaI
0.25µL PstI50 pLacI Maxiprep (A1) 990 1 43.5 5 0.25µL EcoRI
0.25µL SpeI50 sRBS-Lum-dT Maxiprep(B8) 4780 2 (of 1:10 dilution) 42.5 5 0.25µL XbaI
0.25µL PstI50 sRBS-Lum-dT (B7) 4375 2.5 (of 1:10 dilution) 42 5 0.25µL XbaI
0.25µL PstI50 pLacI (D6) 440 2 42.5 5 0.25µL EcoRI
0.25µL SpeI50 sRBS-Lum-dT (G2) 335 3 41.5 5 0.25µL XbaI
0.25µL PstI50 sRBS-Lum-dT (G3) 540 2 42.5 5 0.25µL XbaI
0.25µL PstI50 pSB1T3 25 12.5 7 5 0.25µL EcoRI
0.25µL PstI50 Incubate for 30 minutes at 37oC (Start- 12:10pm; End- 12:40pm)
Heat kill enzymes at 80oC for 20 minutes
Ligation:
In a 10µL final volume, add:- 2µL of sRBS-Lum-dT component
- 2µL of pLacI component
- 2µL of pSB1T3 component
- 1µL of T4 Buffer
- 0.25µL of T4 DNA Ligase
- 2.75µL of MilliQ H2O
Incubate for 30 minutes at room temperature to ligate
Incubate for 20 minutes at 80oC to heat kill
June 3/2010
Carried out protocol described in June 2/2010 - Evening
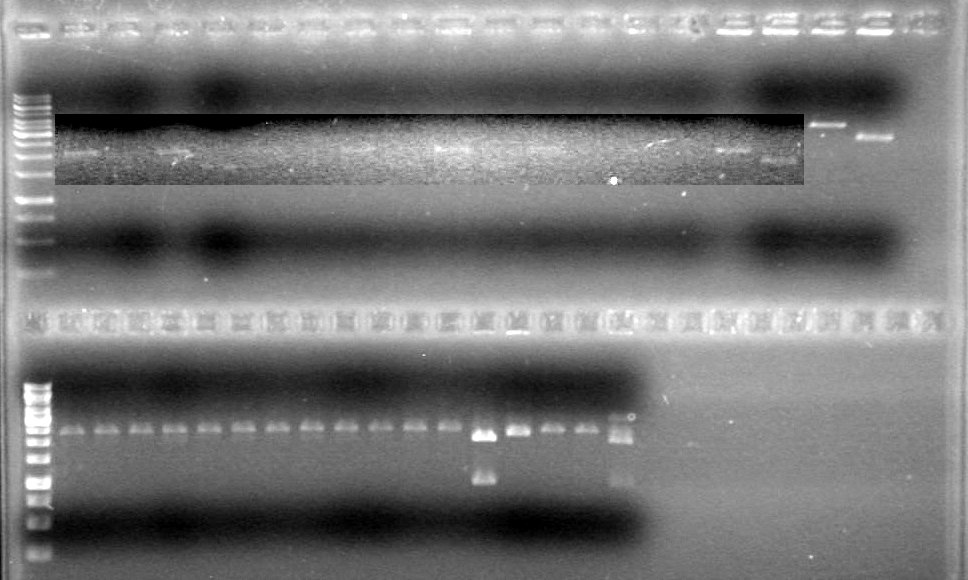
Analyzed results on 1% agarose gel.Load order as follows:
Lane Gel 1
SampleGel 1 Load Gel 2
SampleGel 2 Load 1 1kb Ladder 2µL dye, 2µL ladder
8µL MilliQ H2O1kb Ladder 2µL dye, 2µL ladder
8µL MilliQ H2O2 Restricted
sRBS-Lum-dT (A1)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(A9)+sRBS-Lum-dT(A1)10µL DNA
2µL Dye3 Unrestricted
sRBS-Lum-dT (A1)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(D6)+sRBS-Lum-dT(A1)10µL DNA
2µL Dye4 Restricted
sRBS-Lum-dT (A2)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(A9)+sRBS-Lum-dT(A2)10µL DNA
2µL Dye5 Unrestricted
sRBS-Lum-dT (A2)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(D6)+sRBS-Lum-dT(A2)10µL DNA
2µL Dye6 Restricted
sRBS-Lum-dT (B8)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(A9)+sRBS-Lum-dT(B7)10µL DNA
2µL Dye7 Unrestricted
sRBS-Lum-dT (B8)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(D6)+sRBS-Lum-dT(B7)10µL DNA
2µL Dye8 Restricted
sRBS-Lum-dT (B7)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(A9)+sRBS-Lum-dT(G2)10µL DNA
2µL Dye9 Unrestricted
sRBS-Lum-dT (B7)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(D6)+sRBS-Lum-dT(G2)10µL DNA
2µL Dye10 Restricted
sRBS-Lum-dT (G2)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(D6)+sRBS-Lum-dT(G3)10µL DNA
2µL Dye11 Unrestricted
sRBS-Lum-dT (G2)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(A9)+sRBS-Lum-dT(G3)10µL DNA
2µL Dye12 Restricted
sRBS-Lum-dT (G3)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(D6)+sRBS-Lum-dT(B8)10µL DNA
2µL Dye13 Unrestricted
sRBS-Lum-dT (G3)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(A9)+sRBS-Lum-dT(B8)10µL DNA
2µL Dye14 Restricted
pLacI (A9)10µL DNA
2µL DyeRestricted
rbs-xylE10µL DNA
2µL Dye15 Unrestricted
pLacI (A9)10µL DNA
2µL DyeUnrestricted
rbs-xylE10µL DNA
2µL Dye16 Restricted
pLacI B1 (D6)10µL DNA
2µL DyeRestricted
pSB1T310µL DNA
2µL Dye17 Unrestricted
pLacI B1 (D6)10µL DNA
2µL DyeUnrestricted pSB1T3 10µL DNA
2µL Dye18 Unrestricted
dT10µL DNA
2µL DyepSB1T3 Ligation of:
rbs-xylE+dT10µL DNA
2µL Dye19 Restricted
dT10µL DNA
2µL DyeRan gel at 100V for 90 minutes.
Results:
Conclusions:
June 3/2010 - Evening
Objective: Repeat restriction of pSB1T3 and ligate with pLacI and sRBS-Lum-dT. Previous ligations all used up on gel.
Method:
Ligation:
In a 10µL final volume, add:- 2µL of sRBS-Lum-dT component
- 2µL of pLacI component
- 2µL of pSB1T3 component
- 1µL of T4 Buffer
- 0.25µL of T4 DNA Ligase
- 2.75µL of MilliQ H2O
Incubate for 30 minutes at room temperature to ligate
Incubate for 20 minutes at 80oC to heat kill
Following ligation, transformed using {{Team:Lethbridge/Notebook/Protocols|transformation protocol]]. Plates incubated in 37oC incubator for 44 hours.
Results: Only plate pLacI (D6) + sRBS-Lum-dT (G2) + pSB1T3 grew; had 2 colonies. Control plate did not grow, acidentally plated on tetracycline plate instead of ampicillin (pUC19).
Follow-up: Inoculated 5mL LB media (tetracycline positive) with cells from the transformation plates and incubated at 37oC overnight.
Objective:</br> Ligate pBad-TetR part with fluorescent protein part in pSB1C3 backbone.
- Restrict rbs-xylE wit EcoRI and SpeI (Red Buffer)
 "
"


 ]
]
 ]
]
 ]
]
 ]
]
 ]
]
 ]
]