Team:TU Delft/Modeling/HC regulation
From 2010.igem.org
Lbergwerff (Talk | contribs) (→Hydrocarbon regulated expression system) |
(→Hydrocarbon regulated expression system) |
||
| Line 1: | Line 1: | ||
| - | == Hydrocarbon regulated expression system == | + | {{Team:TU_Delft/frame_check}} |
| + | ==Hydrocarbon regulated expression system== | ||
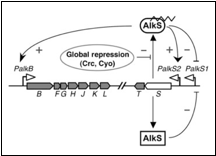
The regulation in ''P. putida'' is shown in the following figure; | The regulation in ''P. putida'' is shown in the following figure; | ||
Revision as of 12:29, 28 September 2010
Hydrocarbon regulated expression system
The regulation in P. putida is shown in the following figure;
There are two operons in this system, the AlkS operon regulated by pAlkS1 and pAlkS2 and the AlkB operon regulated by pAlkB. In reality the AlkB operon contains more genes coding for the degradation pathway, but for simplicity it is just called AlkB.
The pAlkS1 promoter is repressed by any form of AlkS. This promoter has as function maintaining a basal level of AlkS. The pAlkS2 promotor is activated by AlkS bound to hydrocarbons. The function of this promoter is to quickly increase the AlkS concentration in presence of hydrocarbons. The pAlkB promoter behaves as the pAlkS2 promoter and is activated by AlkS bound to hydrocarbons.
The global regulation is done with the protein crc in P. putida. This protein binds to the mRNA of AlkS inhibiting the synthese of AlkS. When substrates preferred over alkanes are present, crc is present. This way if the cell has better options, it will not generate proteins for the hydrocarbon degradation pathway. The mechanism of global regulator cyo is not yet well understood. The level of cyo is dependent on oxygen levels and substrate present.
- link to the experimental part?
Explain the model build-up
- here will be a list with all the assumptions soon, this is still under construction.
Detailed results
Go back
 "
"