Team:TU Delft/Project/solubility/characterization
From 2010.igem.org
(→Characterization of the Solubility Parts) |
|||
| Line 5: | Line 5: | ||
==Emulsifier Assay== | ==Emulsifier Assay== | ||
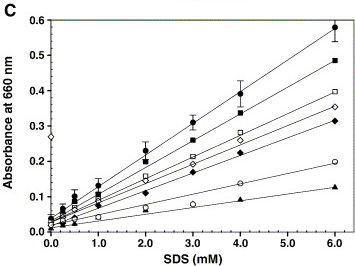
| - | [[Image:TU_Delft_Rajakumari.jpg|thumb| | + | [[Image:TU_Delft_Rajakumari.jpg|thumb|350px|right|Calibration graph showing a linear increase in absorbance at 660 nm for increasing concentrations of SDS (●). Under the standard detergent assay conditions, various components like 200 mM NaCl (□), 2.0 mM CaCl2 (♦), 10% glycerol (■), 100 μg microsomal membranes (open diamond), 0.2 mM Triton X-100 (▲) and 2.5 mM CHAPS (○) were added and the turbidity was measured. Error bars represent the deviation from five independent experiments and each one performed in duplicate. ''Rajakumari et al'' (2006) Biochemical and Biophysical Methods]]We developed a new assay to measure the emulsification caused by AlnA. Assays currently described in literature involve spectrophotometric measurements of turbidity after mixing. Although many articles show nice graphs (see textbox on the right), we were unable to reproduce them. Measuring turbidity turned out to be rather arbitrary. |
| - | [[Image:TU_Delft_sudan_ii_balls_stick.png|thumb|right| | + | [[Image:TU_Delft_sudan_ii_balls_stick.png|thumb|right|350px|Ball-and-stick model of the Sudan II molecule, an orange-red azo dye used for staining of non-polar substances. Source [http://commons.wikimedia.org/wiki/File:Sudan-II-3D-balls.png Wikipedia]]]Instead of a colorless hydrocarbon we choose to use the orange Sudan II dye. |
===Protein Isolation Protocol=== | ===Protein Isolation Protocol=== | ||
| Line 51: | Line 51: | ||
====Calibration and Background==== | ====Calibration and Background==== | ||
| - | [[Image:TU_Delft_emulsification_assay_calibration_curve.jpg|thumb| | + | [[Image:TU_Delft_emulsification_assay_calibration_curve.jpg|thumb|350px|right|Emulsification Assay calibration curve]]The assay was calibrated using SDS to be able to compare it to existing assays. Our measurements are shown in the graph on right. It shows that the assay is very sensible at low SDS concentrations. |
Next, the influence of the culture media was determined. We used LB medium for the initial growth of the cells, and induced the production of AlnA in M9 medium. The background influence of the media on the assay were measured, as shown below. It shows that LB medium already has some emulsification effect, but M9 shows very little influence. | Next, the influence of the culture media was determined. We used LB medium for the initial growth of the cells, and induced the production of AlnA in M9 medium. The background influence of the media on the assay were measured, as shown below. It shows that LB medium already has some emulsification effect, but M9 shows very little influence. | ||
Revision as of 14:34, 8 September 2010
Contents |
Characterization of the Solubility Parts
Emulsifier Production
The alna gene was induced when the culture reached a density of about 108 bacteria per mL. Bacterial and medium samples were taken for sodium dodecyl sulfate (SDS)-gel electrophoresis to monitor AlnA production. The results shown in Fig. ? indicate that expression of AlnA begins ? min after induction and peaks after ? h.
Emulsifier Assay
Protein Isolation Protocol
Materials:
- 25 mM triethanolamine (TEA) buffer, pH 8
- 1% lysozyme in TEA
- 4M urea
- 10% streptomycin in TEA
- Glass beads
- 25 mM Tris buffer (pH 8)
Method:
- Harvest 25 mL bacterial cells by centrifugation at 10.000 rpm for 10 min.
- Collect the cell free supernatant and store it.
- Wash pellet with TEA buffer.
- Freeze pellet in -70 C for 15 min.
- Resuspend pellet 2 mL TEA + 40 ul 1% lysozyme (final concentration 0.02%).
- Incubate for 10 min at RT.
- Disrupt cells with glass beads. Vortex for 10 minutes.
- Centrifuge at 14.000 rpm for 30 min at 4 C (protein is in supernatant).
- Aliquot 1.8 mL supernatant into fresh eppendorf tubes and add 200 ul 10% streptomycin in TEA (final concentration 1%).
- Incubate 10 min at RT.
- Centrifuge at 14.000 rpm for 30 min at 4 C (protein is in pellet).
- Resuspend pellet in 4 M urea.
- Centrifuge at 14.000 rpm for 30 min at 4 C (protein is in supernatant).
Final protein concentrations were measured using Bradford assay.
Emulsification Assay
Materials:
- 25 mM Tris buffer (pH 8)
- 0.1% Sudan II in 50% EtOH
Method:
- Aliquot protein sample in a cuvet.
- Add Tris buffer up to 1 mL.
- Add 20 ul Sudan II.
- Vortex 15 sec at max speed.
- Let sample rest for 1 min.
- Measure absorbance at 493 nm.
Calibration and Background
Next, the influence of the culture media was determined. We used LB medium for the initial growth of the cells, and induced the production of AlnA in M9 medium. The background influence of the media on the assay were measured, as shown below. It shows that LB medium already has some emulsification effect, but M9 shows very little influence.
 "
"