Team:Calgary/23 July 2010
From 2010.igem.org
| (16 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{CalgaryNotebookTemplate| | {{CalgaryNotebookTemplate| | ||
| - | + | Friday July 23, 2010| | |
| - | + | ||
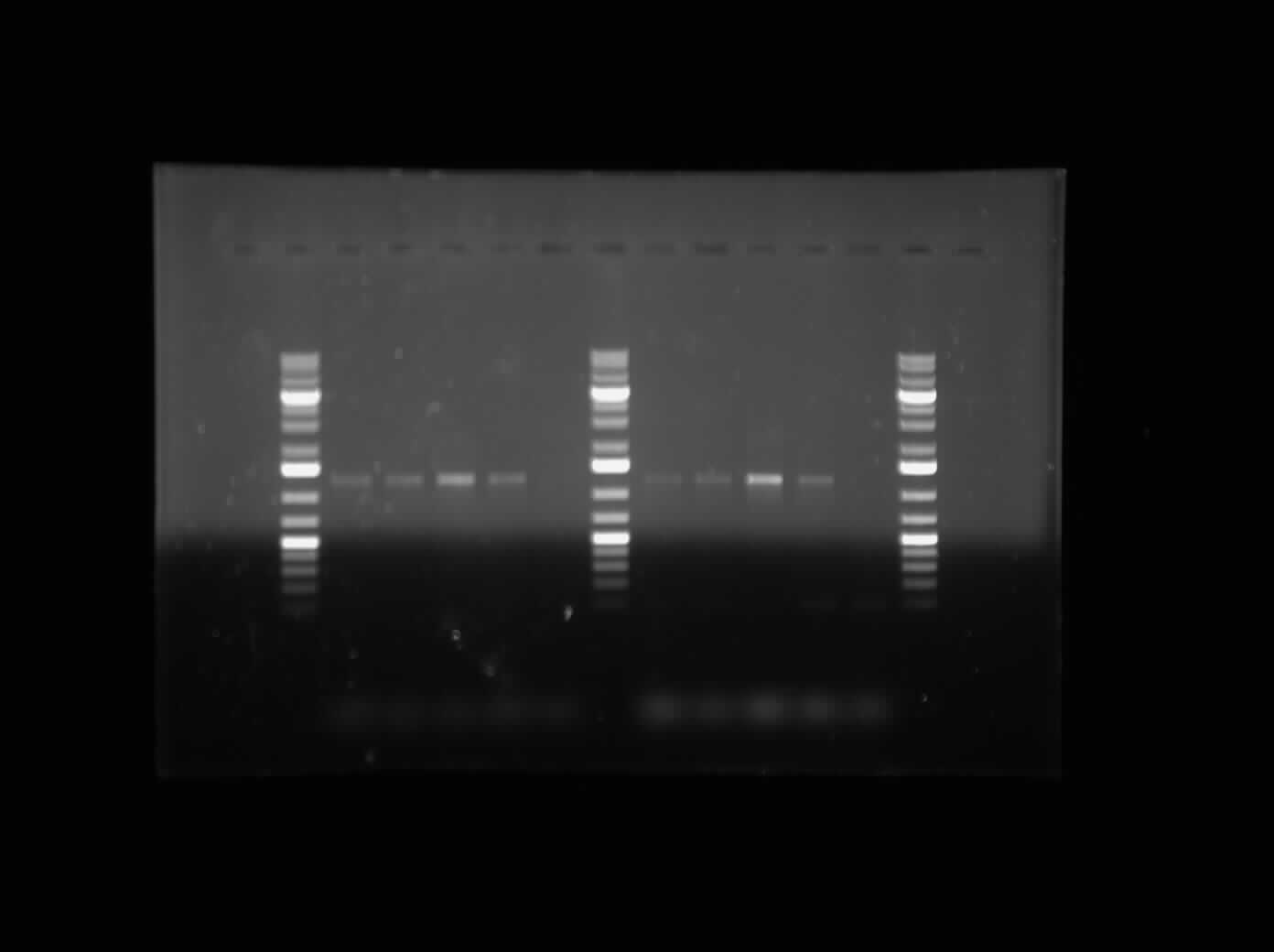
[[Image:07.23.2010. MalEGeneSpec.jpg|thumb|400px|Gel electrophoresis of Raida's PCR products: Mal-E Gene specific primers]] | [[Image:07.23.2010. MalEGeneSpec.jpg|thumb|400px|Gel electrophoresis of Raida's PCR products: Mal-E Gene specific primers]] | ||
| + | |||
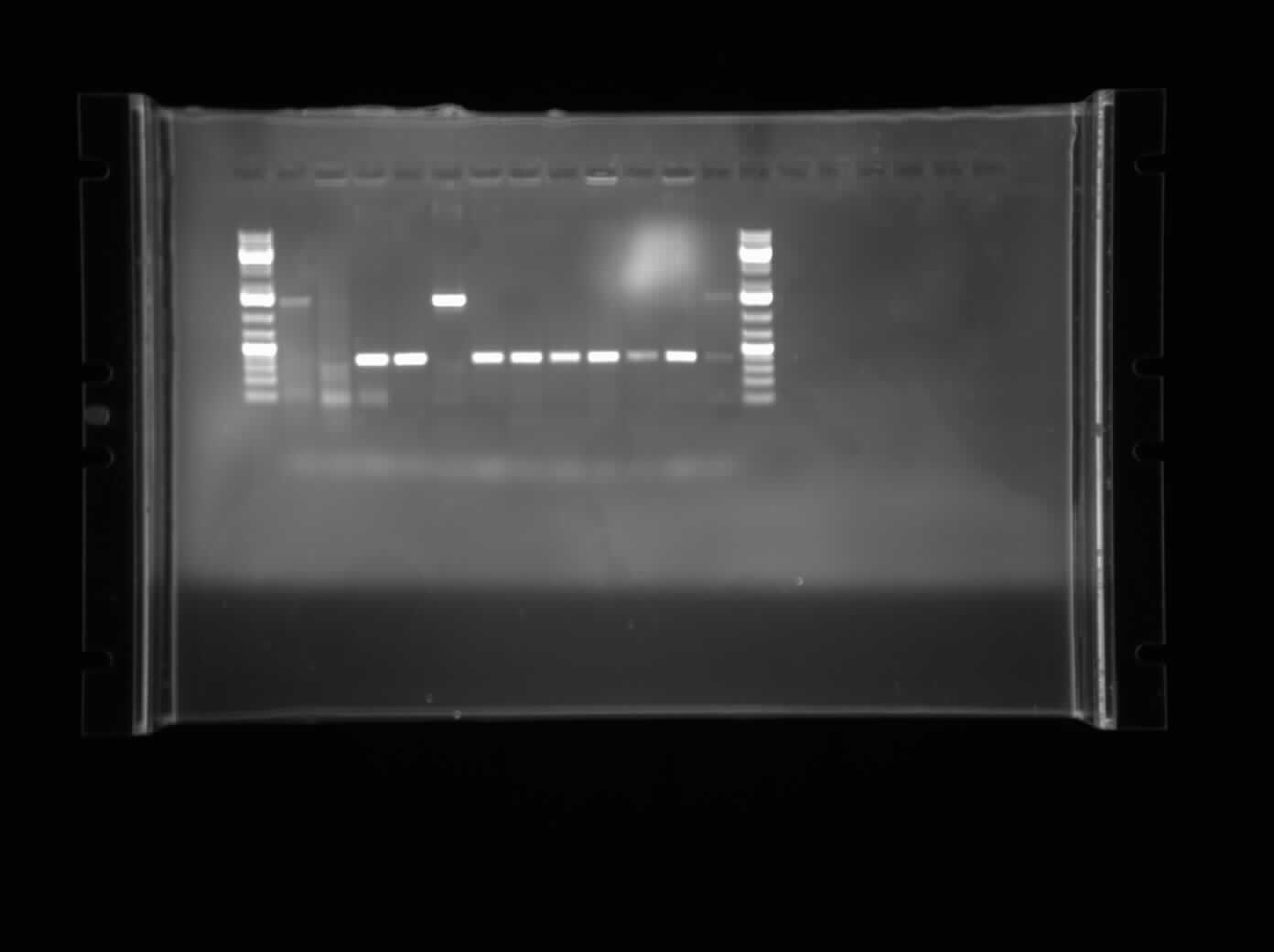
| + | [[Image:07.23.2010. CpxP Gradient PCR.jpg|thumb|400px|Chris's Gradient PCR gel electrophoresis product of the CpxP promoter. The bands were more specific and showed around 400 bp, which is the size of the desired plasmid. A regular PCR and restriction digest will be run to continue to verify.]] | ||
| + | |||
| + | [[Image:07.23.2010.himika-I0500-B0034-PSB1AC3.jpg|thumb|400px|Himika's colony PCR of I0500-B0034 construction in pSB1AC3. The bands were expected to be around 1400 bp and they turned out to be as expected. Lane 2 and 3 will be restreaked, plasmid prepped and sequenced.]] | ||
| + | [[Image:07.23.2010.JeremyK135000+I13504orI13507.jpg|thumb|400px|Jeremy's K135000(CpxR Promoter)+I13507(B0034+E1010+B0015) and I13504 (B0034+E0040+B0015)]] | ||
| + | [[Image:07.23.2010.JeremyK239000+I13504orI13507.jpg|thumb|400px|Jeremy's K139000(DegP promoter)+I13507(B0034+E1010+B0015) and I13504 (B0034+E0040+B0015)]] | ||
<u> Raida </u> | <u> Raida </u> | ||
| - | The first thing I did this morning was run a Gel Electrophoresis of my Mal-E Gene Specific Primer Testing PCR products. I ran it on a 1% gel at 100 V for an hour. Please refer to the image to the side. As it can be seen, the PCR results are positive. All the bands are near the 1200 bp band size, which indicates that it is the Mal-E gene. Furthermore, Lane 7 and Lane 13 show no band because this was the DNA that had the deleted sequence. So the fact that there is no band there is an indication of the fact that the '''MalE primer is functional and only amplified the MalE gene as it was supposed to'''. Further testing will be done to assure that the band shows the MalE Gene. | + | * The first thing I did this morning was run a Gel Electrophoresis of my Mal-E Gene Specific Primer Testing PCR products. I ran it on a 1% gel at 100 V for an hour. Please refer to the image to the side. As it can be seen, the PCR results are positive. All the bands are near the 1200 bp band size, which indicates that it is the Mal-E gene. Furthermore, Lane 7 and Lane 13 show no band because this was the DNA that had the deleted sequence. So the fact that there is no band there is an indication of the fact that the '''MalE primer is functional and only amplified the MalE gene as it was supposed to'''. Further testing will be done to assure that the band shows the MalE Gene. |
| + | * After setting up the PCR I also prepared and ran a 1% gel at 100 V to test the bands of the I0500-B0034. | ||
| + | |||
| + | |||
| + | <u>Himika</u> | ||
| + | |||
| + | Today I ran a colony PCR of 11 of the colonies from the plates last night which was I0500-B0034 plasmid switched into pSB1AC3. Most of the colony PCRs seem to work, although the bands are faint. | ||
| + | Primers used: | ||
| + | |||
| + | *Bbk_CP_F | ||
| + | *Bbk_CP_R | ||
| + | |||
| + | These two primers anneal 100 bp upstream and downstream of the biobrick prefix and suffix. The expected size of the bands was 1400 bp which was exactly what was seen in the gel. I have decided to forward with B1,C1 which are lanes 2 and 3 respectively. | ||
| + | |||
| + | I also restreaked the two colonies on AC plates and left it in the incubator at 20 C. I also made overnight cultures of the same colonies which will be taken out of the shaker on saturday 24 July, 2010. | ||
| + | |||
| + | |||
| + | <u>Dev</u> | ||
| + | |||
| + | This week I worked on the t-shirt designs in Inkscape. The designs are almost finalized. I also helped miniprep the construction of K239000+I13507 today. | ||
| + | |||
| + | |||
| + | <u>Emily</u> | ||
| + | |||
| + | Today I diluted the malE-BBK primers as well as the pRFP-M primers for the site-directed mutagenesis of pRFP. I ran a PCR of malE and malE31 in order to try to Biobrick it. I also ran a PCR of my old construct of I0500-B0034 with the new BBK-CP-F and BBK-CP-R primers. If Himika's I0500-B0034 construct does not give positive reuslts after sequencing, then we may try this one out as a back-up. Raida's PCR of malE and malE31 with gene specific primers as well as the primers that add XbaI and SpeI restriction sites looked really good, so I went ahead and PCR purified the PCR product from the reactions set up with the XbaI/ SpeI primers. Raida also diluted the PCR product that had been run with the gene specific primers and we ran this as a template in two reactions of the malE-BBK PCR. | ||
| + | |||
| + | |||
| + | <u>Chris</u> | ||
| + | |||
| + | Today, I ran an agarose gel electrophoresis of the gradient PCR that was run overnight last night. The bands came up as shown on the right. The bands that appeared were in the range of approximately 400 bp for several of the lanes. This is the projected size of the part of the CpxP promoter as well as the plasmid it was inserted into. After this, I set up a restriction digest with XbaI and PstI which would then be run on an agarose gel electrophoresis to verify the size. Finally, a regular plasmid PCR was set up to determine the size of the part. This will be left to run overnight. Contact was also made with several different companies for funding and possible sponsorships for our project. Several companies that we are contacting have different time zones and thus the times must be coordinated so they are open and we are here. Packages to mail to companies were started but will be finished for next week to mail. | ||
| + | |||
| + | |||
| + | <u>Patrick</u> | ||
| + | |||
| + | I finally found a few things regarding OD<sub>600</sub> and cell concentrations, though there was only one quantitative correlation, and a note stating that the growth curve is mostly linear up until an OD<sub>600</sub> of about 0.6. In a way, this looks promising, though it doesn't tell us ''too'' much. | ||
| + | |||
| + | I will look into potential stresses of protein overproduction on the cell Monday. | ||
| + | |||
| + | <u>Jeremy</u> | ||
| + | I did a 24 well PCR of the K135000+I13507 and I13504 as well as K239000+I13507 and I13504. I did 6 colonies for each notably 3 red colonies for K239000+I13507. The gel that this was run at was a 0.8% agarose set at 90 V and left for ~40 minutes. Constructs were K135000 and K239000 as vectors so in some of the lanes where I13504 and I13504 were not added,the promoter can be seen by itself. The K135000+I13504 PCR only showed K135000 so Alex is redoing the construction. For the K135000+I13507, wells 10, 11, 12, and 14 has promising results however there is some indication that in some of the tubes there is only K135000 as well. For K239000+I13504 wells 4 and 5 has some growth however more experimentation needs to occur before anything can be guaranteed. For K239000+I13507, wells 9-13 all have indication that the construction was successful. From this point, Alex and Dev are continuing the K135000 and K239000 constructions. | ||
| - | |||
}} | }} | ||
Latest revision as of 04:49, 23 August 2010

Friday July 23, 2010
Raida
- The first thing I did this morning was run a Gel Electrophoresis of my Mal-E Gene Specific Primer Testing PCR products. I ran it on a 1% gel at 100 V for an hour. Please refer to the image to the side. As it can be seen, the PCR results are positive. All the bands are near the 1200 bp band size, which indicates that it is the Mal-E gene. Furthermore, Lane 7 and Lane 13 show no band because this was the DNA that had the deleted sequence. So the fact that there is no band there is an indication of the fact that the MalE primer is functional and only amplified the MalE gene as it was supposed to. Further testing will be done to assure that the band shows the MalE Gene.
- After setting up the PCR I also prepared and ran a 1% gel at 100 V to test the bands of the I0500-B0034.
Himika
Today I ran a colony PCR of 11 of the colonies from the plates last night which was I0500-B0034 plasmid switched into pSB1AC3. Most of the colony PCRs seem to work, although the bands are faint. Primers used:
- Bbk_CP_F
- Bbk_CP_R
These two primers anneal 100 bp upstream and downstream of the biobrick prefix and suffix. The expected size of the bands was 1400 bp which was exactly what was seen in the gel. I have decided to forward with B1,C1 which are lanes 2 and 3 respectively.
I also restreaked the two colonies on AC plates and left it in the incubator at 20 C. I also made overnight cultures of the same colonies which will be taken out of the shaker on saturday 24 July, 2010.
Dev
This week I worked on the t-shirt designs in Inkscape. The designs are almost finalized. I also helped miniprep the construction of K239000+I13507 today.
Emily
Today I diluted the malE-BBK primers as well as the pRFP-M primers for the site-directed mutagenesis of pRFP. I ran a PCR of malE and malE31 in order to try to Biobrick it. I also ran a PCR of my old construct of I0500-B0034 with the new BBK-CP-F and BBK-CP-R primers. If Himika's I0500-B0034 construct does not give positive reuslts after sequencing, then we may try this one out as a back-up. Raida's PCR of malE and malE31 with gene specific primers as well as the primers that add XbaI and SpeI restriction sites looked really good, so I went ahead and PCR purified the PCR product from the reactions set up with the XbaI/ SpeI primers. Raida also diluted the PCR product that had been run with the gene specific primers and we ran this as a template in two reactions of the malE-BBK PCR.
Chris
Today, I ran an agarose gel electrophoresis of the gradient PCR that was run overnight last night. The bands came up as shown on the right. The bands that appeared were in the range of approximately 400 bp for several of the lanes. This is the projected size of the part of the CpxP promoter as well as the plasmid it was inserted into. After this, I set up a restriction digest with XbaI and PstI which would then be run on an agarose gel electrophoresis to verify the size. Finally, a regular plasmid PCR was set up to determine the size of the part. This will be left to run overnight. Contact was also made with several different companies for funding and possible sponsorships for our project. Several companies that we are contacting have different time zones and thus the times must be coordinated so they are open and we are here. Packages to mail to companies were started but will be finished for next week to mail.
Patrick
I finally found a few things regarding OD600 and cell concentrations, though there was only one quantitative correlation, and a note stating that the growth curve is mostly linear up until an OD600 of about 0.6. In a way, this looks promising, though it doesn't tell us too much.
I will look into potential stresses of protein overproduction on the cell Monday.
Jeremy
I did a 24 well PCR of the K135000+I13507 and I13504 as well as K239000+I13507 and I13504. I did 6 colonies for each notably 3 red colonies for K239000+I13507. The gel that this was run at was a 0.8% agarose set at 90 V and left for ~40 minutes. Constructs were K135000 and K239000 as vectors so in some of the lanes where I13504 and I13504 were not added,the promoter can be seen by itself. The K135000+I13504 PCR only showed K135000 so Alex is redoing the construction. For the K135000+I13507, wells 10, 11, 12, and 14 has promising results however there is some indication that in some of the tubes there is only K135000 as well. For K239000+I13504 wells 4 and 5 has some growth however more experimentation needs to occur before anything can be guaranteed. For K239000+I13507, wells 9-13 all have indication that the construction was successful. From this point, Alex and Dev are continuing the K135000 and K239000 constructions.
 "
"