Team:SDU-Denmark/labnotes5
From 2010.igem.org
(→Lab notes (8/9 - 8/15)) |
(→Lab notes (8/9 - 8/15)) |
||
| Line 206: | Line 206: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | After this the standarts and samples were measured on a UV-vis spetrometer. | + | After this the standarts and samples were measured on a UV-vis spetrometer.<br> |
| + | [[Image:Team-SDU-denmarkBeta-carotene.jpg |400px]] | ||
=== PCR of NinaB (again) === | === PCR of NinaB (again) === | ||
Start date: 13/8<br> | Start date: 13/8<br> | ||
Revision as of 11:32, 18 August 2010
Lab notes (8/9 - 8/15)
Contents |
Group: Photosensor
pfu PCR of B0015 and PCR purification
date: 17/8
Protocols: CP1.1 and GFX easy protocol
Notes:
2 uL PCR product of B0015 (no. 43 white) was used as template for each PCR reaction.3 PCR reactions were prepared.
Premix for 4 PCR reactions:
| 25uL | pfu bf. + MgSO4 |
| 7,5uL | dNTP's |
| 7,5uL | VF2 |
| 7,5uL | VR |
| 190uL | H20 |
| 2uL | pfu Polymerase enzyme |
48uL premix is distrubuted into each PCR tube. PCR tubes are marked B0015.A-C
PCR program:
| Start | 95C | 3min |
| Denaturating | 95C | 2min |
| Annealing | 55C | 30s |
| Elongation | 72C | 45s |
| Go to | 2 | 29x |
| End | 72C | 2min |
| Hold | 4C |
5uL of PCR sample is loaded onto a 2% agarose gel. Generuler 100bp DNA ladder (blue) is used as marker.
Results:
Analysis:
PCR product is OK and B0015 DNA is purified from the PCR product according to protocol. DNA is eluted in 20uL H2O. Purified samples are stored in the jumblebox (hotchpotch), marked as B0015-1 and B0015-2.
Group: Retinal
Characterization of Beta-carotene
Start date: 10/8
Methods: ON, sonication, UV-vis spectroscopy
Protocols: CC.1.1[1]
Colony PCR on transformants using ninaB fwd and rw primers
Date: 10/8
Done by: Christian & Tommy
Methods: ON
protocos:
Notes:
ON was made from 4 colonies:
Top 10 - on insert -> OD = 0,008 (100 x diluted)
Top 10 - with K274210 biobrick insert -> OD = 0,011 (100 x diluted)
MG1655 - on insert -> OD = 0,020 (100 x diluted)
Mg1655 - with K274210 biobrick insert -> OD = 0,017 (100 x diluted)
ON was made in 110 ml LB media. colonies with K274210 biobrick insert was grown in media containing ampicillin.
alle the ON cultures was grown for 20 hours at 37 °C.
After 16 hours, ml of the ON colonies was transferred in to 110 LB media and grown for 4 hours to the exponential phase:
Top 10 - on insert -> OD = 0,049 (100 x diluted)
Top 10 - with K274210 biobrick insert -> OD = 0,044 (100 x diluted)
MG1655 - on insert -> OD = 0,007 (100 x diluted)
Mg1655 - with K274210 biobrick insert -> OD = 0,009 (100 x diluted)
Colonies with K274210 biobrick insert was grown in media containing ampicillin.
Harvesting, sonication and UV-vis spektroscopy
Date: 11/8
Done by: Christian & Tommy
Methods: Cell harvesting, sonication and UV-vis messurements
protocos: none
Notes:
100 mL Cell cultures was centrifuged for 5 min at 14000 RPM. the supernantant was discarded and cells were resuspended in 5 mL acetone (99,9%), except the top 10 with the K274210 biobrick insert wicht was resuspended in 10 mL acetone (suorce of error.). The resuspended cells were sonicated for 5 min. the samples were spun down and the supernantant were transferrede to new tubes, cell bebri was discarded.
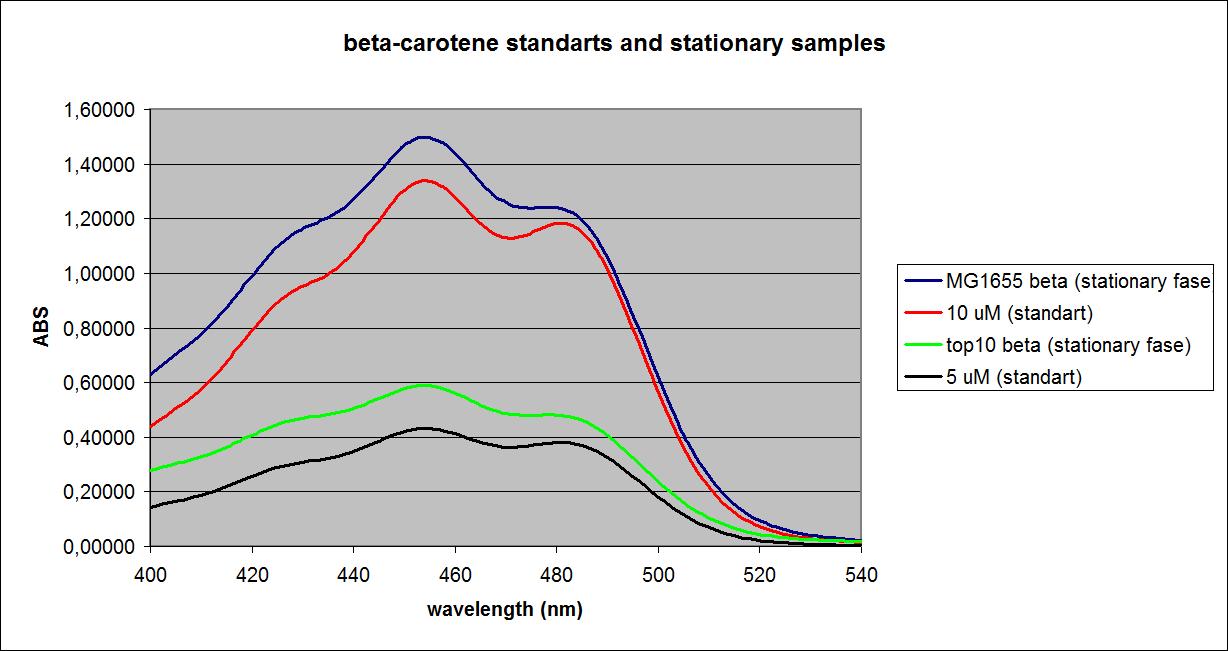
A standart curve was made from pure Beta-carotene,
The samples and the standarts were first measured at a fixed wavelengt of 456 nm.
The standarts:
| Concentration | Absorbance |
| 1 mM | 4,000 |
| 100 µM | 2,260 |
| 50 µM | 4,000 |
| 25 µM | 0,155 |
| 10 µM | 0,893 |
| 5 µM | 0,440 |
| 1 µM | 0,075 |
| 100 nM | 0,015 |
| 10 nM | 0,038 |
| 1 nM | 0,005 |
| 100 pM | 0,024 |
The samples:
| Top 10 cells (Absorbance) | MG1655 E. coli mutant (Absorbance) | |
| Stationary phase control | 0,034 | 0,024 |
| Stationary phase with K274210 biobrick insert | 0,319 | 1,549 |
| Expotential phase control | 0,020 | 0,024 |
| Expotential phase with K274210 biobrick insert | 0,034 | 0,033 |
After this the standarts and samples were measured on a UV-vis spetrometer.
PCR of NinaB (again)
Start date: 13/8
Methods: Restriction digest, PCR, gel
Protocols: RD1.1 [2], CC.1.1[3]
Colony PCR on transformants using ninaB fwd and rw primers
Date: 13/8
Done by: Marie & Tommy
Methods: Restriction digest, PCR, gel
protocos: RD1.1, CC.1.1
Notes:
Restirction digest was performed with EcoRI according to protocol. (gel was run on protocol).
PCR was run on the product (No gel purification).
The following PCR program was used:
| PCR | Temp. (C) | Time |
| Start | 95 | 2 min |
| Denaturation | 95 | 45 sec |
| Annealing | 49 | 30 sec |
| Elongation | 72 | 4 min |
| Denaturing | 95 | 45 sec |
| Annealing | 69 | 30 sec |
| Elongation | 72 | 4 min |
| End | 72 | 5 min |
| Hold | 4 | indef. |
gel was run on the PCR product.
Miniprep of POT2 with NinaB insert
Start date: 13/8
Methods: Miniprep, Restriction digest, gel
Protocols: MP1.2 [4], RD1.1 [5].
ON of Top 10 E. coli cells with POT2 with NinaB
Date: 16/8
Done by: Marie & Tommy
Methods:
protocos:
Notes:
100 mL ON culture was made, cells were grown at 37 C in LB media with Chloramphenicol.
ON of Top 10 E. coli cells with POT2 with NinaB
Date: 17/8
Done by: Marie & Tommy
Methods: Miniprep, Restriction digest, gel
Protocols: MP1.2 [6], RD1.1 [7].
Notes:
50 mL of elution buffer was used. DNA concentrations after pooling were measured on NanoDrop to 206,2 ng/microL.
gel was run on produckt, showing recent miniprep compared to old miniprep produckt, showing uneven lengths. subsequent wa performed.
Restriction digest on miniprep produckt (w. EcoRI)
Date: 17/8
Done by: Marie & Tommy
Methods: Restriction digest, gel
Protocols: MP1.2 [8], gel.
Notes:
Due to the lack of old sample, restriction digest was performed using only 3,5 microL of miniprep produckt. 1,5 microL of H2O was added insted. EcoRI was used
 "
"