Team:TU Delft/10 August 2010 content
From 2010.igem.org
(Difference between revisions)
(→Tolerance and Sensing) |
(→Tolerance and Sensing) |
||
| Line 63: | Line 63: | ||
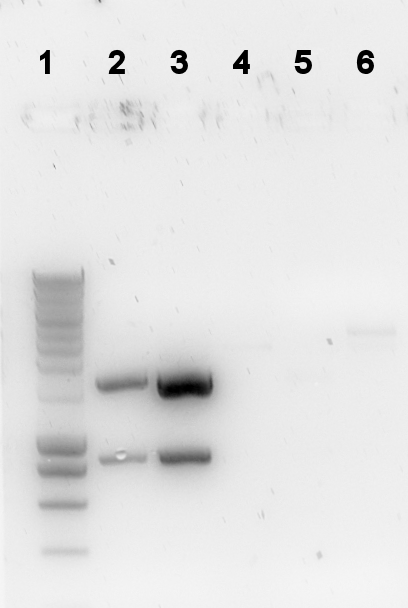
The digestion was checked on a gel: | The digestion was checked on a gel: | ||
| - | [[Image:TU_Delft_2010-08-10_digestion_p7.png]] | + | |
| + | [[Image:TU_Delft_2010-08-10_digestion_p7.png|thumb]] | ||
Lane description | Lane description | ||
Revision as of 12:24, 16 August 2010
Contents |
Lab work
Tolerance and Sensing
A new attempt to construct parts 304 (PalkS12 + B0032), 327(P(caif) + B0032) and 405 (402 + 403).
Digestion, Ligation, Transformation
1 microgram of each plasmid was digested:
| # | Sample | Enzyme 1 | Enzyme 2 | Enzyme 3 | Buffer | BSA | Needed fragment |
| 1 | PalkS12A | EcoRI | SpeI | ScaI | 2 (Biolabs) | ✓ | ‘E - PalkS12 - S’ |
| 2 | P(caif)A | EcoRI | SpeI | ScaI | 2 (Biolabs) | ✓ | ‘E - P(caif) - S’ |
| 3 | B0032 | EcoRI | XbaI | 2 (Biolabs) | ✓ | ‘X - B0032 - E’ | |
| 4 | 402: J61101 - PhPFDalpha | EcoRI | SpeI | ScaI | 2 (Biolabs) | ✓ | ‘E - J61101 - PhPFDalpha - S’ |
| 5 | 403: J61101 - PhPFDbeta | EcoRI | XbaI | 2 (Biolabs) | ✓ | ‘E - J61101 - PhPFDbeta - S’ |
The digestion was checked on a gel:
Lane description
| # | Description | Expected size (bp) | OK? |
| L | Smartladder (5μl) | n/a | n/a |
| 1 | PalkS cut | 106, 861, 1188 | no |
| 2 | P(caif) cut | 74, 861, 118 | no |
| 4 | 402 cut | 543, 548, 1508 | no |
| 5 | 403 cut | 2439, 58 | vague |
Alkane degradation
All of yesterday's plates contained colonies. To check whether they contain the correct insert we will do a colony PCR.

Lane description
| # | Description | Primers | Expected length (bp) | ✓ | ✗ |
| L | SmartLadder | n/a | n/a | n/a | n/a |
| 1-5 | 013A | G00100 + G00101 | 1929 | none | all |
| 6-10 | 013K | G00100 + G00101 | 1929 | 9 | 6,7,8,10 |
| 11-15 | 020A | G00100 + G00101 | 2482 | none | all |
| L | SmartLadder | n/a | n/a | n/a | n/a |
On the gel it looks like lane 9, containing 013K, has the correct length. We will do a plasmid isolation tomorrow. Unfortunately none of the colonies of 020A seemed to be correct. We will check some more colonies with a colony PCR overnight.
 "
"