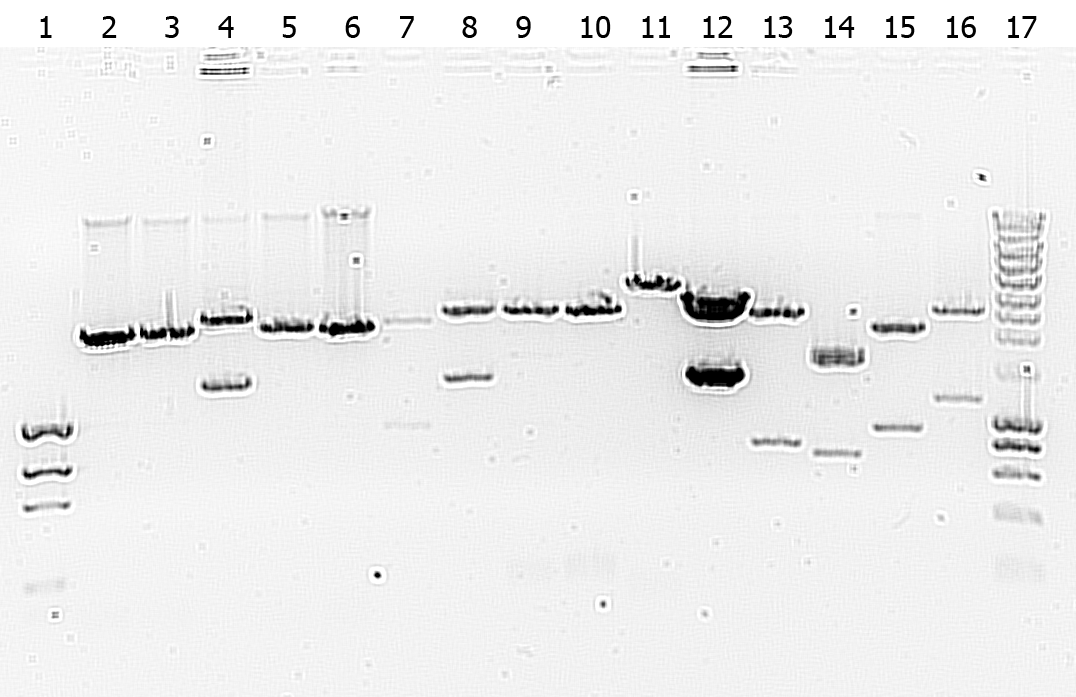Team:TU Delft/19 July 2010 content
From 2010.igem.org
(→Lab work) |
(→Characterization of Anderson RBS sequences) |
||
| (24 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | =Forbidden word= | |
Today we chose a word that we were not allowed to say all day. The word was pipet. It turned out to be hard not to use this word. All kind of funny definitions were used to avoid the word pipet. Still some persons accidentally said the word and have to do the dishes tomorrow. | Today we chose a word that we were not allowed to say all day. The word was pipet. It turned out to be hard not to use this word. All kind of funny definitions were used to avoid the word pipet. Still some persons accidentally said the word and have to do the dishes tomorrow. | ||
=Lab work= | =Lab work= | ||
| - | + | ==Ordered DNA + Solvent Tolerance and Hydrocarbon Sensing== | |
| - | The frozen pellet of [https://2010.igem.org/Team:TU_Delft/ | + | The frozen pellet of [https://2010.igem.org/Team:TU_Delft#page=pages/blog&blog=16_July_2010 last week] were isolated using [[Team:TU_Delft/protocols/birnboim_plasmid_isolation|Birnboim protocol]]. |
| - | The following plasmid concentrations were obtained: | + | The following plasmid concentrations were obtained using the nanodrop: |
{| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
|'''BioBrick''' | |'''BioBrick''' | ||
| Line 13: | Line 13: | ||
|'''Concentration (ng/μL)''' | |'''Concentration (ng/μL)''' | ||
|- | |- | ||
| - | | | + | |K398328T |
| - | | | + | |AlkS and B0015 |
| - | | | + | |145.01 |
|- | |- | ||
| - | | | + | |K398407T |
| - | | | + | |PhPFDa and B0032 |
| - | | | + | |37.8 |
|- | |- | ||
| - | | | + | |Control |
| - | | | + | |pSB1T3 and B0015 |
| - | | | + | |71.65 |
|- | |- | ||
| - | | | + | |K398300C |
| - | | | + | |AlkS |
| - | | | + | |24.54 |
|- | |- | ||
| - | | | + | |Control |
| - | | | + | |pSB1C3 |
| - | | | + | |93.86 |
|- | |- | ||
| - | | | + | |K398200C |
| - | | | + | |AlnA |
| - | | | + | |4.23 |
|- | |- | ||
| - | | | + | |K398000C |
| - | | | + | |LadA |
| - | | | + | |148.14 |
| + | |- | ||
| + | |K398002C | ||
| + | |RubA3 | ||
| + | |119.85 | ||
| + | |- | ||
| + | |K398201C | ||
| + | |OprG | ||
| + | |161.28 | ||
| + | |- | ||
| + | |K398400C | ||
| + | |PhPFDa | ||
| + | |163.33 | ||
|} | |} | ||
| - | Because not all [https://2010.igem.org/Team:TU_Delft/ | + | Because not all [https://2010.igem.org/Team:TU_Delft#page=pages/blog&blog=14_July_2010 ligation mixes] resulted in transformants were repeated the [[Team:TU_Delft/protocols/transformation|transformation]] with the overnight ligated reactions. |
| - | + | ==Alkane degradation== | |
Biobricks in production: K398007T, K398008T, K398009T, K398010T, K398016T, K398017T & K398018T | Biobricks in production: K398007T, K398008T, K398009T, K398010T, K398016T, K398017T & K398018T | ||
| - | + | ====>Digestion==== | |
[[Team:TU_Delft/protocols/restriction_enzyme_digestion| Digestion reactions]]: | [[Team:TU_Delft/protocols/restriction_enzyme_digestion| Digestion reactions]]: | ||
{| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
|'''#''' | |'''#''' | ||
| - | |''' | + | |'''Sample''' |
| - | |''' | + | |'''Enzyme 1''' |
| + | |'''Enzyme 2''' | ||
| + | |'''Buffer''' | ||
| + | |'''BSA''' | ||
|'''Needed fragment''' | |'''Needed fragment''' | ||
|- | |- | ||
|1 | |1 | ||
| - | |2 μg J61100 | + | |2 μg J61100 |
| - | | | + | |EcoRI |
| - | | | + | |SpeI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘E–J61100–S’ | ||
|- | |- | ||
|2 | |2 | ||
| - | |2 μg J61100 | + | |2 μg J61100 |
| - | | | + | |EcoRI |
| - | | | + | |SpeI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘E–J61100–S’ | ||
|- | |- | ||
|3 | |3 | ||
| - | |1 μg rubR | + | |1 μg rubR |
| - | | | + | |EcoRI |
| - | | | + | |SpeI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘E–rubR–S’ | ||
|- | |- | ||
|4 | |4 | ||
| - | |1 μg J61101 | + | |1 μg J61101 |
| - | | | + | |EcoRI |
| - | | | + | |SpeI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘E–J61101–S’ | ||
|- | |- | ||
|5 | |5 | ||
| - | |1 μg J61107 | + | |1 μg J61107 |
| - | | | + | |EcoRI |
| - | | | + | |SpeI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘E–J61107–S’ | ||
|- | |- | ||
|6 | |6 | ||
| - | |1 μg alkB2 | + | |1 μg alkB2 |
| - | | | + | |Xbal |
| - | | | + | |PstI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘X–alkB2–P’ | ||
|- | |- | ||
|7 | |7 | ||
| - | |1 μg rubA3 | + | |1 μg rubA3 |
| - | | | + | |Xbal |
| - | | | + | |PstI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘X–rubA3–P’ | ||
|- | |- | ||
|8 | |8 | ||
| - | |1 μg rubA4 | + | |1 μg rubA4 |
| - | | | + | |Xbal |
| - | | | + | |PstI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘X–rubA4–P’ | ||
|- | |- | ||
|9 | |9 | ||
| - | |1 μg B0015 | + | |1 μg B0015 |
| - | | | + | |Xbal |
| - | | | + | |PstI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘X–B0015–P’ | ||
|- | |- | ||
|10 | |10 | ||
| - | |1 μg ladA | + | |1 μg ladA |
| - | | | + | |Xbal |
| - | | | + | |PstI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘X–ladA–P’ | ||
|- | |- | ||
|11 | |11 | ||
| - | |1 μg ADH | + | |1 μg ADH |
| - | | | + | |Xbal |
| - | |‘X | + | |PstI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘X-ADH–P’ | ||
|- | |- | ||
|12 | |12 | ||
| - | |1 μg ALDH | + | |1 μg ALDH |
| - | | | + | |Xbal |
| - | | | + | |PstI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘X–ALDH–P’ | ||
|- | |- | ||
|13 | |13 | ||
| - | |3 μg pSB1T3 | + | |3 μg pSB1T3 |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘E–linear pSB1T3–P’ | ||
|} | |} | ||
| Line 125: | Line 179: | ||
Results of the digestion on [[Team:TU_Delft/protocols/agarose_gel|1% agarose gel]] | Results of the digestion on [[Team:TU_Delft/protocols/agarose_gel|1% agarose gel]] | ||
| - | [[Image:TUDelft_19jul2010_digestion.png|500px|thumb|left| | + | [[Image:TUDelft_19jul2010_digestion.png|500px|thumb|left|1% agarose of digestion check. Gel runned at 100V for 1 hour. Of all samples 5 μL was loaded with 1 μL loadingbuffer. 5 μL was loaded of marker]] |
| - | + | ||
Lane description: | Lane description: | ||
| Line 133: | Line 186: | ||
|'''Description''' | |'''Description''' | ||
|'''Expected Length (bp)''' | |'''Expected Length (bp)''' | ||
| + | |'''Status''' | ||
|'''Remarks''' | |'''Remarks''' | ||
|- | |- | ||
|1 | |1 | ||
| - | |Easy load marker | + | |Easy load marker |
| + | |n/a | ||
|n/a | |n/a | ||
| | | | ||
|- | |- | ||
|2 | |2 | ||
| - | | | + | |J61100 + EcoRI + SpeI |
| - | |78 | + | |78, 2056 |
| + | |? | ||
|Not visible (too small) | |Not visible (too small) | ||
|- | |- | ||
|3 | |3 | ||
| - | | | + | |J61100 + EcoRI + SpeI |
| - | |78 | + | |78, 2056 |
| + | |? | ||
|Not visible (too small) | |Not visible (too small) | ||
|- | |- | ||
|4 | |4 | ||
| - | | | + | |rubR + EcoRI + SpeI |
| - | |1227 | + | |1227, 2326 |
| - | | | + | |<font color=limegreen>✓</font> |
| + | | | ||
|- | |- | ||
|5 | |5 | ||
| - | | | + | |J61101 + EcoRI + SpeI |
| - | |78 | + | |78, 2056 |
| + | |? | ||
|Not visible (too small) | |Not visible (too small) | ||
|- | |- | ||
|6 | |6 | ||
| - | | | + | |J61107 + EcoRI + SpeI |
| - | |78 | + | |78, 2056 |
| + | |? | ||
|Not visible (too small) | |Not visible (too small) | ||
|- | |- | ||
|7 | |7 | ||
| - | | | + | |pSB1A2-J23100 + SpeI + PstI |
|2110 | |2110 | ||
| - | | | + | |<font color=limegreen>✓</font> |
| + | |low concentration | ||
|- | |- | ||
|8 | |8 | ||
| - | | | + | |alkB2 + XbaI + PstI |
| - | | | + | |1255, 2411 |
| - | | | + | |<font color=limegreen>✓</font> |
| + | | | ||
|- | |- | ||
|9 | |9 | ||
| - | | | + | |rubA3 + XbaI + PstI |
| - | |198 | + | |198, 2409 |
| + | |? | ||
|Not visible on picture, but was vaguely seen | |Not visible on picture, but was vaguely seen | ||
|- | |- | ||
|10 | |10 | ||
| - | | | + | |rubA4 + XbaI + PstI |
| - | |207 | + | |207, 2409 |
| + | |? | ||
|Not visible on picture, but was vaguely seen | |Not visible on picture, but was vaguely seen | ||
|- | |- | ||
|11 | |11 | ||
| - | | | + | |B0015 + XbaI + PstI |
| - | |155 | + | |155, 3163 |
| + | |? | ||
|Not visible (too small) | |Not visible (too small) | ||
|- | |- | ||
|12 | |12 | ||
| - | | | + | |ladA + XbaI + PstI |
| - | |1350 | + | |1350, 2915 |
| - | | | + | |<font color=limegreen>✓</font> |
| + | | | ||
|- | |- | ||
|13 | |13 | ||
| - | | | + | |ADH + XbaI + PstI |
| - | | | + | |769, 2411 |
| - | | | + | |<font color=limegreen>✓</font> |
| + | | | ||
|- | |- | ||
|14 | |14 | ||
| - | | | + | |ALDH + XbaI + PstI |
| - | |1521 | + | |1521, 1618, 705 |
| + | |<font color=limegreen>✓</font> | ||
|Originally in a Kan backbone which gives an extra fragment of 800 bp when cut, rest of the backbone and the fragment both ~ 1500, so overlap | |Originally in a Kan backbone which gives an extra fragment of 800 bp when cut, rest of the backbone and the fragment both ~ 1500, so overlap | ||
|- | |- | ||
|15 | |15 | ||
| - | | | + | |E0240 + XbaI + PstI |
| - | | | + | |902, 2053 |
| - | | | + | |<font color=limegreen>✓</font> |
| + | | | ||
|- | |- | ||
|16 | |16 | ||
| - | | | + | |pSB1T3 + EcoRI + PstI |
| - | |2422 | + | |2422, 1350 |
| - | | | + | |<font color=limegreen>✓</font> |
| + | | | ||
|- | |- | ||
|17 | |17 | ||
| - | | | + | |[https://2010.igem.org/Image:TU_Delft_SmartLadder.jpg SmartLadder] |
| + | |n/a | ||
|n/a | |n/a | ||
| | | | ||
|} | |} | ||
| - | + | ====Ligation==== | |
| - | + | ||
| - | + | ||
The digestion products were [[Team:TU_Delft/protocols/ligation|ligated]] over night: | The digestion products were [[Team:TU_Delft/protocols/ligation|ligated]] over night: | ||
| Line 294: | Line 363: | ||
To all samples with an end volume of 25μL (and 25.8μL), 2.5 μL Ligase buffer was added. For those with an end volume of 30μL (indicated by an *) 3.0μL Ligase buffer was added. | To all samples with an end volume of 25μL (and 25.8μL), 2.5 μL Ligase buffer was added. For those with an end volume of 30μL (indicated by an *) 3.0μL Ligase buffer was added. | ||
| - | + | ==Emulsifier== | |
Since last weeks attempt to construct the inducible protomer R0011 with RBS B0032 failed, we decided to take a different approach. Instead of doing the standard 2 parts assembly, we amplify the promoter by PCR digest it and ligate in the plasmid that contains the RBS which has only be cut open at the left side. By doing so we do not risk losing the super small DNA fragments like the RBS. The problem is that we cannot use antibiotics selection. So we need to determine that by colony PCR and sequencing. | Since last weeks attempt to construct the inducible protomer R0011 with RBS B0032 failed, we decided to take a different approach. Instead of doing the standard 2 parts assembly, we amplify the promoter by PCR digest it and ligate in the plasmid that contains the RBS which has only be cut open at the left side. By doing so we do not risk losing the super small DNA fragments like the RBS. The problem is that we cannot use antibiotics selection. So we need to determine that by colony PCR and sequencing. | ||
| - | + | ====PCR Amplification==== | |
R0011 was [[Team:TU_Delft/protocols/PCR|amplified]] with the universal primers G00100 and G00101. The product was put on [[Team:TU_Delft/protocols/agarose_gel|1% agarose gel]]: | R0011 was [[Team:TU_Delft/protocols/PCR|amplified]] with the universal primers G00100 and G00101. The product was put on [[Team:TU_Delft/protocols/agarose_gel|1% agarose gel]]: | ||
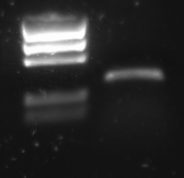
| - | [[Image:TU_Delft_P1_2010-07-19_PCR_Product_promotor.png|thumb|right|1% agarose | + | [[Image:TU_Delft_P1_2010-07-19_PCR_Product_promotor.png|thumb|right|1% agarose of PCR. Gel runned at 100 V for 1 hour. Of all samples 10 μL was loaded with 2 μL loadingbuffer. 5 μL was loaded of marker]] |
Lane description: | Lane description: | ||
| Line 308: | Line 377: | ||
|'''#''' | |'''#''' | ||
|'''Description''' | |'''Description''' | ||
| + | |'''Expected length (bp)''' | ||
| + | |'''Primers''' | ||
| + | |'''Status''' | ||
| + | |'''Remarks''' | ||
|- | |- | ||
| - | | | + | |M1 |
| - | |BioRad EZ Ladder | + | |BioRad EZ Ladder |
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
|- | |- | ||
| - | | | + | |1 |
| - | |PCR Product of R0011 | + | |PCR Product of R0011 |
| + | |293 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=limegreen>✓</font> | ||
| + | |R0011 itself is just 55 bp, but the flanking primer regions are about 100 bp each | ||
|} | |} | ||
The PCR band is about 300 bps long. R0011 itself is just 55 bp, but the flanking primer regions are about 100 bp each. | The PCR band is about 300 bps long. R0011 itself is just 55 bp, but the flanking primer regions are about 100 bp each. | ||
| - | + | ====Digestion==== | |
The PCR product of promotor R0011 was cut with EcoRI and SpeI and the RBS containing plasmid has been cut open with EcoRI and Xbal for 2.5 hours at 37 °C. This deviates from the standard biobrick assembly, thus is not completely as described in our [[Team:TU_Delft/protocols/restriction_enzyme_digestion|digestion protocol]]. | The PCR product of promotor R0011 was cut with EcoRI and SpeI and the RBS containing plasmid has been cut open with EcoRI and Xbal for 2.5 hours at 37 °C. This deviates from the standard biobrick assembly, thus is not completely as described in our [[Team:TU_Delft/protocols/restriction_enzyme_digestion|digestion protocol]]. | ||
| Line 324: | Line 405: | ||
{| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
|'''#''' | |'''#''' | ||
| - | |''' | + | |'''Sample''' |
| - | |''' | + | |'''Enzyme 1''' |
| + | |'''Enzyme 2''' | ||
| + | |'''Buffer''' | ||
| + | |'''BSA''' | ||
|'''Needed fragment''' | |'''Needed fragment''' | ||
|- | |- | ||
|1 | |1 | ||
| - | |1 μg B0032 | + | |1 μg B0032 |
| - | | | + | |EcoRI |
| - | | | + | |Xbal |
| + | |2 (BioLabs) | ||
| + | | | ||
| + | |‘X–B0032 pSB1A2–E’ | ||
|- | |- | ||
|2 | |2 | ||
| - | |30 μL PCR product of R0011 | + | |30 μL PCR product of R0011 |
| - | | | + | |EcoRI |
| - | |‘E - R0011 - S’ | + | |SpeI |
| + | |2 (BioLabs) | ||
| + | | | ||
| + | |‘E-R0011-S’ | ||
|} | |} | ||
| - | + | <h4>Ligation</h4> | |
The digestion products were [[Team:TU_Delft/protocols/ligation|ligated]] over night: | The digestion products were [[Team:TU_Delft/protocols/ligation|ligated]] over night: | ||
| Line 345: | Line 435: | ||
{| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
|'''#''' | |'''#''' | ||
| - | |''' | + | |'''BioBrick''' |
| + | |'''Fragment 1''' | ||
| + | |'''Fragment 2''' | ||
|- | |- | ||
|1 | |1 | ||
| - | |5 μL | + | |K398202 |
| + | |5 μL ‘X–B0032 pSB1A2–E’ | ||
| + | |10 μL ‘E-R0011-S’ | ||
|} | |} | ||
| - | Hopefully this will lead to 'E - X - R0011 - B0032 - S - P' in the B0032 plasmid backbone with Ampicillin resistance marker. | + | Hopefully this will lead to 'E-X-R0011-B0032-S-P' in the B0032 plasmid backbone with Ampicillin resistance marker. |
| - | + | ==Competent cells== | |
Top10 and DH5α cells were cultivated for making competent cells | Top10 and DH5α cells were cultivated for making competent cells | ||
==Characterization of Anderson RBS sequences== | ==Characterization of Anderson RBS sequences== | ||
<h5>Assembly of reference construct</h5> | <h5>Assembly of reference construct</h5> | ||
| - | [https://2010.igem.org/Team:TU_Delft#/blog | + | [https://2010.igem.org/Team:TU_Delft#page=pages/blog&blog=16_July_2010 Friday's] ligation products, ligation control and digestion control were [https://2010.igem.org/Team:TU_Delft/protocols/transformation transformed] into chemically competent Top10 cells and incubated on ampicillin plates. |
| - | + | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | |
| + | |'''#''' | ||
| + | |'''BioBrick''' | ||
| + | |'''Plasmid''' | ||
| + | |- | ||
| + | |T1 | ||
| + | |K081005-I13401 | ||
| + | |pSB1A2 | ||
| + | |- | ||
| + | |T2 | ||
| + | |K081005-I13401 | ||
| + | |pSB1A2 | ||
| + | |- | ||
| + | |T3 | ||
| + | |I13401 (ligation control) | ||
| + | |pSB1AK2 | ||
| + | |- | ||
| + | |T4 | ||
| + | |'E-pSB1A2-I13401-X' (digest control) | ||
| + | |pSB1AK2 | ||
| + | |- | ||
| + | |T5 | ||
| + | |MilliQ | ||
| + | |'''-''' | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | Similarly a third attempt was made at transforming K081005, this time into commercial chemically competent cells. | ||
{| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
|'''#''' | |'''#''' | ||
| - | |''' | + | |'''BioBrick''' |
| - | |''' | + | |'''Plasmid''' |
| - | |''' | + | |- |
| + | |T6 | ||
| + | |K081005 | ||
| + | |pSB1A2 | ||
| + | |} | ||
| + | |||
| + | In order to continue progress made on the fourth method, described in the blog of [https://2010.igem.org/Team:TU_Delft#page=pages/blog&blog=14_July_2010 July 14th] for the assembly of the reference construct J23100-B0032-E0040-B0015, the following [[Team:TU_Delft/protocols/restriction_enzyme_digestion_40| Digestions]] were performed: | ||
| + | |||
| + | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Sample''' | ||
| + | |'''Enzyme 1''' | ||
| + | |'''Enzyme 2''' | ||
| + | |'''Buffer''' | ||
| + | |'''BSA''' | ||
| + | |'''Needed fragment''' | ||
|- | |- | ||
|1 | |1 | ||
| - | |1 μg J23100 | + | |1 μg J23100 |
| - | | | + | |SpeI |
| - | | | + | |PstI |
| + | |2 (BioLabs) | ||
| + | | | ||
| + | |‘S–J23100 in J61002–P’ | ||
|- | |- | ||
|2 | |2 | ||
| - | |1 μg E0240 | + | |1 μg E0240 |
| - | | | + | |XbaI |
| - | | | + | |PstI |
| + | |2 (BioLabs) | ||
| + | | | ||
| + | |‘X–E0240–P’ | ||
|} | |} | ||
After enzyme inactivation, the digestion products were set for [https://2010.igem.org/Team:TU_Delft/protocols/ligation ligation] overnight. | After enzyme inactivation, the digestion products were set for [https://2010.igem.org/Team:TU_Delft/protocols/ligation ligation] overnight. | ||
| - | + | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | |
| + | |'''#''' | ||
| + | |'''BioBrick''' | ||
| + | |'''Fragment 1''' | ||
| + | |'''Fragment 2''' | ||
| + | |'''Recipient vector''' | ||
| + | |'''Final volume''' | ||
| + | |- | ||
| + | |1 | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K173000 K173000] | ||
| + | |10.5 μL ‘X-E0240-P’ | ||
| + | |'''-''' | ||
| + | |4 μL ‘S-pSB1A2-J23100-P’ | ||
| + | |25 μL | ||
| + | |- | ||
| + | |} | ||
| - | + | To all samples with an end volume of 25 μL, 2.5 μL ligase buffer was added. | |
| - | < | + | In the likelihood that the abovementioned method will not yield desired transformants, a [[Team:TU_Delft/protocols/PCR|PCR amplification]] of E0240 was performed in three-fold in accordance with method 3. The resulting product over 1% agarose gel can be seen below. The PCR amplification product was subsequently digested using XbaI and PstI for later insertion into previously digested (SpeI and PstI) J23100(in J61002). |
| + | |||
| + | [[Team:TU_Delft/protocols/agarose_gel |1% Agarose gel]] of E0240 PCR product: | ||
| + | [[Image:TU_Delft_2010_PCR_19-07_Thias.png|200px|thumb|left|1% agarose of PCR. Gel runned at 100V for 1 hour. Of all samples 5 μL was loaded with 1 μL loadingbuffer. 5 μL was loaded of marker.]] | ||
| + | |||
| + | Lane descriptions: | ||
| + | {|style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Description''' | ||
| + | |'''Expected Length (bp)''' | ||
| + | |'''Primers''' | ||
| + | |'''Status''' | ||
| + | |'''Remarks''' | ||
| + | |- | ||
| + | |1 | ||
| + | |SmartLadder marker | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
| + | |- | ||
| + | |2 | ||
| + | |PCR product of E0240 #1 | ||
| + | |1114 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=limegreen>✓</font> | ||
| + | | | ||
| + | |- | ||
| + | |3 | ||
| + | |PCR product of E0240 #2 | ||
| + | |1114 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=limegreen>✓</font> | ||
| + | | | ||
| + | |- | ||
| + | |4 | ||
| + | |PCR product of E0240 #3 | ||
| + | |1114 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=limegreen>✓</font> | ||
| + | | | ||
| + | |- | ||
| + | |} | ||
Latest revision as of 15:01, 12 August 2010
Contents |
Forbidden word
Today we chose a word that we were not allowed to say all day. The word was pipet. It turned out to be hard not to use this word. All kind of funny definitions were used to avoid the word pipet. Still some persons accidentally said the word and have to do the dishes tomorrow.
Lab work
Ordered DNA + Solvent Tolerance and Hydrocarbon Sensing
The frozen pellet of last week were isolated using Birnboim protocol.
The following plasmid concentrations were obtained using the nanodrop:
| BioBrick | Composed of | Concentration (ng/μL) |
| K398328T | AlkS and B0015 | 145.01 |
| K398407T | PhPFDa and B0032 | 37.8 |
| Control | pSB1T3 and B0015 | 71.65 |
| K398300C | AlkS | 24.54 |
| Control | pSB1C3 | 93.86 |
| K398200C | AlnA | 4.23 |
| K398000C | LadA | 148.14 |
| K398002C | RubA3 | 119.85 |
| K398201C | OprG | 161.28 |
| K398400C | PhPFDa | 163.33 |
Because not all ligation mixes resulted in transformants were repeated the transformation with the overnight ligated reactions.
Alkane degradation
Biobricks in production: K398007T, K398008T, K398009T, K398010T, K398016T, K398017T & K398018T
>Digestion
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA | Needed fragment |
| 1 | 2 μg J61100 | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–J61100–S’ |
| 2 | 2 μg J61100 | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–J61100–S’ |
| 3 | 1 μg rubR | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–rubR–S’ |
| 4 | 1 μg J61101 | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–J61101–S’ |
| 5 | 1 μg J61107 | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–J61107–S’ |
| 6 | 1 μg alkB2 | Xbal | PstI | 2 (BioLabs) | ✓ | ‘X–alkB2–P’ |
| 7 | 1 μg rubA3 | Xbal | PstI | 2 (BioLabs) | ✓ | ‘X–rubA3–P’ |
| 8 | 1 μg rubA4 | Xbal | PstI | 2 (BioLabs) | ✓ | ‘X–rubA4–P’ |
| 9 | 1 μg B0015 | Xbal | PstI | 2 (BioLabs) | ✓ | ‘X–B0015–P’ |
| 10 | 1 μg ladA | Xbal | PstI | 2 (BioLabs) | ✓ | ‘X–ladA–P’ |
| 11 | 1 μg ADH | Xbal | PstI | 2 (BioLabs) | ✓ | ‘X-ADH–P’ |
| 12 | 1 μg ALDH | Xbal | PstI | 2 (BioLabs) | ✓ | ‘X–ALDH–P’ |
| 13 | 3 μg pSB1T3 | EcoRI | PstI | 2 (BioLabs) | ✓ | ‘E–linear pSB1T3–P’ |
Results of the digestion on 1% agarose gel
Lane description:
| # | Description | Expected Length (bp) | Status | Remarks |
| 1 | Easy load marker | n/a | n/a | |
| 2 | J61100 + EcoRI + SpeI | 78, 2056 | ? | Not visible (too small) |
| 3 | J61100 + EcoRI + SpeI | 78, 2056 | ? | Not visible (too small) |
| 4 | rubR + EcoRI + SpeI | 1227, 2326 | ✓ | |
| 5 | J61101 + EcoRI + SpeI | 78, 2056 | ? | Not visible (too small) |
| 6 | J61107 + EcoRI + SpeI | 78, 2056 | ? | Not visible (too small) |
| 7 | pSB1A2-J23100 + SpeI + PstI | 2110 | ✓ | low concentration |
| 8 | alkB2 + XbaI + PstI | 1255, 2411 | ✓ | |
| 9 | rubA3 + XbaI + PstI | 198, 2409 | ? | Not visible on picture, but was vaguely seen |
| 10 | rubA4 + XbaI + PstI | 207, 2409 | ? | Not visible on picture, but was vaguely seen |
| 11 | B0015 + XbaI + PstI | 155, 3163 | ? | Not visible (too small) |
| 12 | ladA + XbaI + PstI | 1350, 2915 | ✓ | |
| 13 | ADH + XbaI + PstI | 769, 2411 | ✓ | |
| 14 | ALDH + XbaI + PstI | 1521, 1618, 705 | ✓ | Originally in a Kan backbone which gives an extra fragment of 800 bp when cut, rest of the backbone and the fragment both ~ 1500, so overlap |
| 15 | E0240 + XbaI + PstI | 902, 2053 | ✓ | |
| 16 | pSB1T3 + EcoRI + PstI | 2422, 1350 | ✓ | |
| 17 | SmartLadder | n/a | n/a |
Ligation
The digestion products were ligated over night:
| # | BioBrick | Fragment 1 | Fragment 2 | Recipient vector | Final volume |
| 1 | K398007T | 6 μL ‘E-J61100-S’ | 12 μL ‘X-alkB2-P’ | 1.3 μL ‘E-pSB1T3-P’ | 25 μL |
| 2 | K398008T | 6 μL ‘E-J61100-S’ | 11 μL ‘X-rubA3-P’ | 1.3 μL ‘E-pSB1T3-P’ | 25 μL |
| 3 | K398009T | 6 μL ‘E-J61100-S’ | 12.5 μL ‘X-rubA4-P’ | 1.3 μL ‘E-pSB1T3-P’ | 25 μL |
| 4 | K398010T | 10 μL ‘E-rubR-S’ | 11 μL ‘X-B0015-P’ | 1.3 μL ‘E-pSB1T3-P’ | 25.8 μL |
| 5 | K398016T | 6 μL ‘E-J61100-S’ | 12 μL ‘X-ladA-P’ | 1.3 μL ‘E-pSB1T3-P’ | 25 μL |
| 6 | K398017T | 12.5 μL ‘E-J61101-S’ | 11 μL ‘X-ADH-P’ | 1.3 μL ‘E-pSB1T3-P’ | 30 μL* |
| 7 | K398018T | 12.5 μL ‘E-J61107-S’ | 11.5 μL ‘X-ALDH-P’ | 1.3 μL ‘E-pSB1T3-P’ | 30 μL* |
| 8 | n/a | 10.5 μL ‘X-E0240-P’ | n/a | 4 μL ‘S-pSB1A2-J23100-P’ | 25 μL |
To all samples with an end volume of 25μL (and 25.8μL), 2.5 μL Ligase buffer was added. For those with an end volume of 30μL (indicated by an *) 3.0μL Ligase buffer was added.
Emulsifier
Since last weeks attempt to construct the inducible protomer R0011 with RBS B0032 failed, we decided to take a different approach. Instead of doing the standard 2 parts assembly, we amplify the promoter by PCR digest it and ligate in the plasmid that contains the RBS which has only be cut open at the left side. By doing so we do not risk losing the super small DNA fragments like the RBS. The problem is that we cannot use antibiotics selection. So we need to determine that by colony PCR and sequencing.
PCR Amplification
R0011 was amplified with the universal primers G00100 and G00101. The product was put on 1% agarose gel:
Lane description:
| # | Description | Expected length (bp) | Primers | Status | Remarks |
| M1 | BioRad EZ Ladder | n/a | n/a | n/a | |
| 1 | PCR Product of R0011 | 293 | G00100 + G00101 | ✓ | R0011 itself is just 55 bp, but the flanking primer regions are about 100 bp each |
The PCR band is about 300 bps long. R0011 itself is just 55 bp, but the flanking primer regions are about 100 bp each.
Digestion
The PCR product of promotor R0011 was cut with EcoRI and SpeI and the RBS containing plasmid has been cut open with EcoRI and Xbal for 2.5 hours at 37 °C. This deviates from the standard biobrick assembly, thus is not completely as described in our digestion protocol.
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA | Needed fragment |
| 1 | 1 μg B0032 | EcoRI | Xbal | 2 (BioLabs) | ‘X–B0032 pSB1A2–E’ | |
| 2 | 30 μL PCR product of R0011 | EcoRI | SpeI | 2 (BioLabs) | ‘E-R0011-S’ |
Ligation
The digestion products were ligated over night:
| # | BioBrick | Fragment 1 | Fragment 2 |
| 1 | K398202 | 5 μL ‘X–B0032 pSB1A2–E’ | 10 μL ‘E-R0011-S’ |
Hopefully this will lead to 'E-X-R0011-B0032-S-P' in the B0032 plasmid backbone with Ampicillin resistance marker.
Competent cells
Top10 and DH5α cells were cultivated for making competent cells
Characterization of Anderson RBS sequences
Assembly of reference construct
Friday's ligation products, ligation control and digestion control were transformed into chemically competent Top10 cells and incubated on ampicillin plates.
| # | BioBrick | Plasmid |
| T1 | K081005-I13401 | pSB1A2 |
| T2 | K081005-I13401 | pSB1A2 |
| T3 | I13401 (ligation control) | pSB1AK2 |
| T4 | 'E-pSB1A2-I13401-X' (digest control) | pSB1AK2 |
| T5 | MilliQ | - |
Similarly a third attempt was made at transforming K081005, this time into commercial chemically competent cells.
| # | BioBrick | Plasmid |
| T6 | K081005 | pSB1A2 |
In order to continue progress made on the fourth method, described in the blog of July 14th for the assembly of the reference construct J23100-B0032-E0040-B0015, the following Digestions were performed:
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA | Needed fragment |
| 1 | 1 μg J23100 | SpeI | PstI | 2 (BioLabs) | ‘S–J23100 in J61002–P’ | |
| 2 | 1 μg E0240 | XbaI | PstI | 2 (BioLabs) | ‘X–E0240–P’ |
After enzyme inactivation, the digestion products were set for ligation overnight.
| # | BioBrick | Fragment 1 | Fragment 2 | Recipient vector | Final volume |
| 1 | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K173000 K173000] | 10.5 μL ‘X-E0240-P’ | - | 4 μL ‘S-pSB1A2-J23100-P’ | 25 μL |
To all samples with an end volume of 25 μL, 2.5 μL ligase buffer was added.
In the likelihood that the abovementioned method will not yield desired transformants, a PCR amplification of E0240 was performed in three-fold in accordance with method 3. The resulting product over 1% agarose gel can be seen below. The PCR amplification product was subsequently digested using XbaI and PstI for later insertion into previously digested (SpeI and PstI) J23100(in J61002).
1% Agarose gel of E0240 PCR product:
Lane descriptions:
| # | Description | Expected Length (bp) | Primers | Status | Remarks |
| 1 | SmartLadder marker | n/a | n/a | n/a | |
| 2 | PCR product of E0240 #1 | 1114 | G00100 + G00101 | ✓ | |
| 3 | PCR product of E0240 #2 | 1114 | G00100 + G00101 | ✓ | |
| 4 | PCR product of E0240 #3 | 1114 | G00100 + G00101 | ✓ |
 "
"