Team:Newcastle/7 July 2010
From 2010.igem.org
(Difference between revisions)
PhilipHall (Talk | contribs) |
PhilipHall (Talk | contribs) |
||
| Line 1: | Line 1: | ||
{{Team:Newcastle/mainbanner}} | {{Team:Newcastle/mainbanner}} | ||
| + | |||
| + | ==Chromosomal prep== | ||
===Results=== | ===Results=== | ||
| Line 8: | Line 10: | ||
===Conclusion=== | ===Conclusion=== | ||
The aim of this whole experiment was to extract genomic DNA from ''B. subtilis'' strain ATCC 6633. In order to test whether we had extracted the DNA, we first used PCR to amplify and then used Gel Electrophoresis to compare the bands of ''ara'' genes to the hundred bps ladder.It worked! | The aim of this whole experiment was to extract genomic DNA from ''B. subtilis'' strain ATCC 6633. In order to test whether we had extracted the DNA, we first used PCR to amplify and then used Gel Electrophoresis to compare the bands of ''ara'' genes to the hundred bps ladder.It worked! | ||
| + | |||
| + | ==LacI BioBrick Construction== | ||
| + | |||
| + | ===Aims=== | ||
| + | * To use PCR to extract ''lacI'' (promoter, ribosome-binding site (RBS) & coding sequence (CDS)) from plasmid pMutin4 and ligate into vector pSB1AT3 in front of red fluorescent protein (RFP). | ||
| + | |||
| + | ===Materials=== | ||
| + | * PCR prduct | ||
| + | * Restriction enzymes EcoR1 and Spe1 | ||
| + | |||
| + | ===Protocol=== | ||
| + | * ''lacI'' PCR product is purified. | ||
| + | * Purified ''lacI'' is digested with EcoR1 and Spe1. | ||
| + | |||
| + | ===Inference=== | ||
| + | * The purified PCR product is cut to give it sticky ends complementary to the digested pSB1AT3. | ||
| + | {{Team:Newcastle/footer}} | ||
| + | |||
| + | |||
{{Team:Newcastle/footer}} | {{Team:Newcastle/footer}} | ||
Revision as of 11:01, 4 August 2010

| |||||||||||||
| |||||||||||||
Contents |
Chromosomal prep
Results
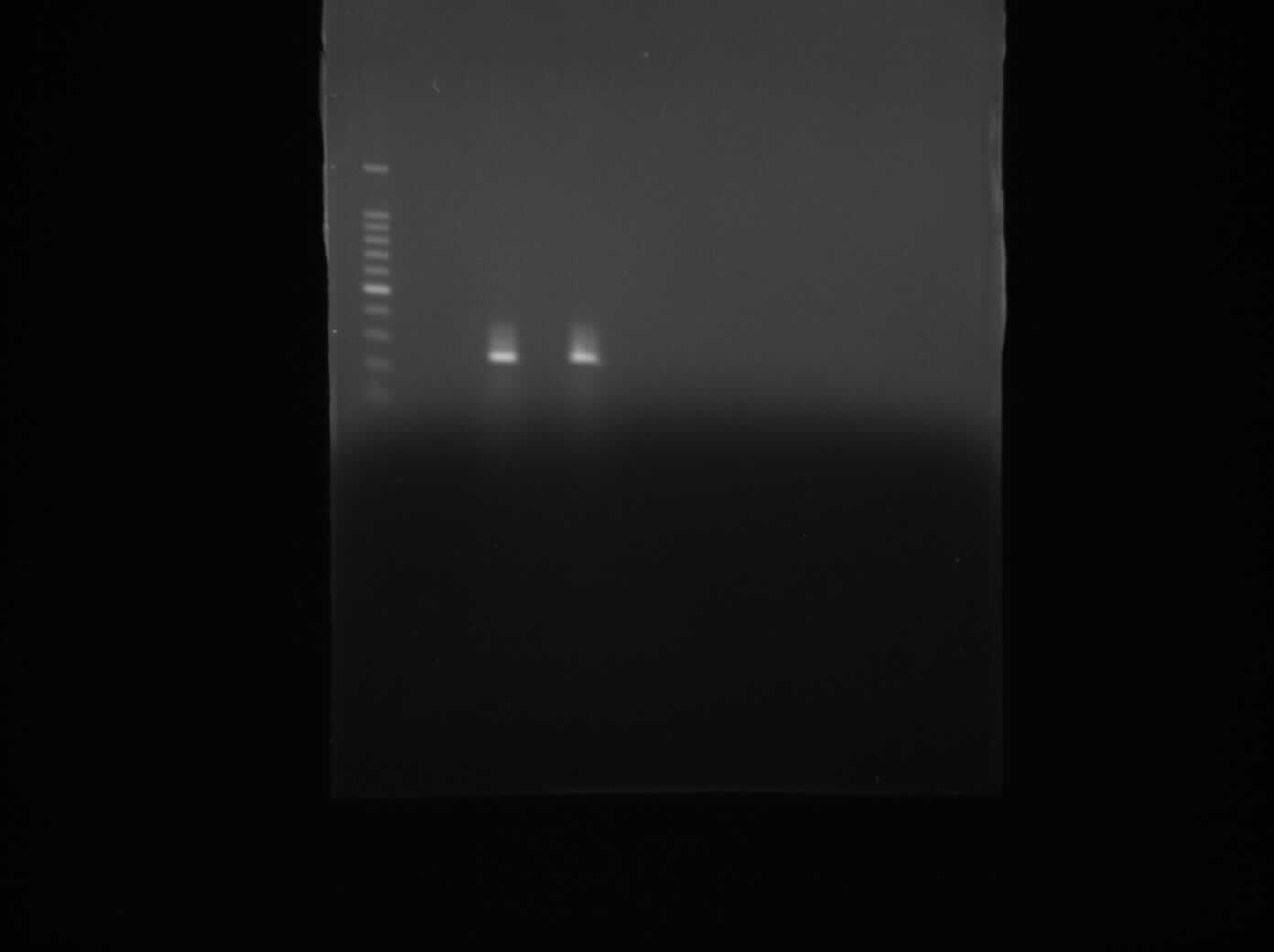
We used Gel Electrophoresis to test whether we have the DNA we wanted. As we already know, the ara gene is about 300bps. We used hundred bp DNA ladder as a guidance for our bands. The two bands produced were from two separate ara cultures and they indeed show the bands in the same region, i.e. 300bps.
Conclusion
The aim of this whole experiment was to extract genomic DNA from B. subtilis strain ATCC 6633. In order to test whether we had extracted the DNA, we first used PCR to amplify and then used Gel Electrophoresis to compare the bands of ara genes to the hundred bps ladder.It worked!
LacI BioBrick Construction
Aims
- To use PCR to extract lacI (promoter, ribosome-binding site (RBS) & coding sequence (CDS)) from plasmid pMutin4 and ligate into vector pSB1AT3 in front of red fluorescent protein (RFP).
Materials
- PCR prduct
- Restriction enzymes EcoR1 and Spe1
Protocol
- lacI PCR product is purified.
- Purified lacI is digested with EcoR1 and Spe1.
Inference
- The purified PCR product is cut to give it sticky ends complementary to the digested pSB1AT3.
 
|
<center>
 
|
 "
"