Team:TU Delft/21 July 2010 content
From 2010.igem.org
(Difference between revisions)
(→Alkane degradation) |
|||
| Line 1: | Line 1: | ||
| + | =Lab work= | ||
| + | |||
| + | ==Ordered DNA== | ||
| + | The transformations of [https://2010.igem.org/Team:TU_Delft#/blog?blog=19_July_2010 19 July] containing the different ligations gave colonies (~5 per plate). We picked 5 colonies per plate and performed a [[Team:TU_Delft/protocols/Colony_PCR|colony PCR]]. | ||
| + | |||
==Alkane degradation== | ==Alkane degradation== | ||
Unfortunately there were no transformants on [https://2010.igem.org/Team:TU_Delft#/blog?blog=20_July_2010 yesterday's] plates. This is most likely due to the fact that the ligation didn't work with the small pieces of DNA of the RBSs. Next plan is to cut open the RBS plasmid without removing the RBS (with SpeI and PstI) and insert the gene in this plasmid. We will try this tomorrow. | Unfortunately there were no transformants on [https://2010.igem.org/Team:TU_Delft#/blog?blog=20_July_2010 yesterday's] plates. This is most likely due to the fact that the ligation didn't work with the small pieces of DNA of the RBSs. Next plan is to cut open the RBS plasmid without removing the RBS (with SpeI and PstI) and insert the gene in this plasmid. We will try this tomorrow. | ||
==Emulsifier== | ==Emulsifier== | ||
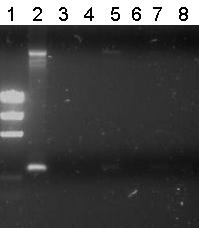
| - | [[Image:TU_Delft_PCR_Pieter_21_07_01.jpg|thumb|right|2% agarose | + | [[Image:TU_Delft_PCR_Pieter_21_07_01.jpg|thumb|right|2% agarose of colony PCR. Gel runned at 100V for 1 hour. Of all samples 10 μL + 2 μL loadingbuffer was loaded. 5 μL was loaded of marker]] There were small colonies on the plates. Pieter picked seven and performed [[Team:TU_Delft/protocols/Colony_PCR|colony PCR]] on them. |
| - | + | Lane Description | |
{| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
|'''#''' | |'''#''' | ||
|'''Description''' | |'''Description''' | ||
|'''Expected lenght (bp)''' | |'''Expected lenght (bp)''' | ||
| + | |'''Primer''' | ||
| + | |'''Status''' | ||
| + | |'''Remarks''' | ||
| + | |- | ||
| + | |M1 | ||
| + | |EZ Ladder | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
|- | |- | ||
|1 | |1 | ||
| - | | | + | |B0032 (control) |
| + | |220 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=limegreen>✔</font> | ||
| | | | ||
|- | |- | ||
|2 | |2 | ||
| - | |B0032 | + | |Transformant #1 of ligation mix R0011-B0032 |
| - | | | + | |300 |
| + | |G00100 + G00101 | ||
| + | |<font color=red>✖</font> | ||
| + | | | ||
|- | |- | ||
|3 | |3 | ||
| - | | | + | |Transformant #2 of ligation mix R0011-B0032 |
|300 | |300 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=red>✖</font> | ||
| + | | | ||
|- | |- | ||
|4 | |4 | ||
| - | | | + | |Transformant #3 of ligation mix R0011-B0032 |
|300 | |300 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=red>✖</font> | ||
| + | | | ||
|- | |- | ||
|5 | |5 | ||
| - | | | + | |Transformant #4 of ligation mix R0011-B0032 |
|300 | |300 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=red>✖</font> | ||
| + | | | ||
|- | |- | ||
|6 | |6 | ||
| - | | | + | |Transformant #5 of ligation mix R0011-B0032 |
|300 | |300 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=red>✖</font> | ||
| + | | | ||
|- | |- | ||
|7 | |7 | ||
| - | | | + | |Transformant #6 of ligation mix R0011-B0032 |
|300 | |300 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=red>✖</font> | ||
| + | | | ||
|- | |- | ||
|8 | |8 | ||
| - | | | + | |Transformant #7 of ligation mix R0011-B0032 |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|300 | |300 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=red>✖</font> | ||
| + | | | ||
|} | |} | ||
| - | + | Unfortunately all other lanes were empty or the same as the negative control. So from this gel we conclude that the ligation has failed. | |
Revision as of 19:14, 1 August 2010
Contents |
Lab work
Ordered DNA
The transformations of 19 July containing the different ligations gave colonies (~5 per plate). We picked 5 colonies per plate and performed a colony PCR.
Alkane degradation
Unfortunately there were no transformants on yesterday's plates. This is most likely due to the fact that the ligation didn't work with the small pieces of DNA of the RBSs. Next plan is to cut open the RBS plasmid without removing the RBS (with SpeI and PstI) and insert the gene in this plasmid. We will try this tomorrow.
Emulsifier
There were small colonies on the plates. Pieter picked seven and performed colony PCR on them.
Lane Description
| # | Description | Expected lenght (bp) | Primer | Status | Remarks |
| M1 | EZ Ladder | n/a | n/a | n/a | |
| 1 | B0032 (control) | 220 | G00100 + G00101 | ✔ | |
| 2 | Transformant #1 of ligation mix R0011-B0032 | 300 | G00100 + G00101 | ✖ | |
| 3 | Transformant #2 of ligation mix R0011-B0032 | 300 | G00100 + G00101 | ✖ | |
| 4 | Transformant #3 of ligation mix R0011-B0032 | 300 | G00100 + G00101 | ✖ | |
| 5 | Transformant #4 of ligation mix R0011-B0032 | 300 | G00100 + G00101 | ✖ | |
| 6 | Transformant #5 of ligation mix R0011-B0032 | 300 | G00100 + G00101 | ✖ | |
| 7 | Transformant #6 of ligation mix R0011-B0032 | 300 | G00100 + G00101 | ✖ | |
| 8 | Transformant #7 of ligation mix R0011-B0032 | 300 | G00100 + G00101 | ✖ |
Unfortunately all other lanes were empty or the same as the negative control. So from this gel we conclude that the ligation has failed.
 "
"