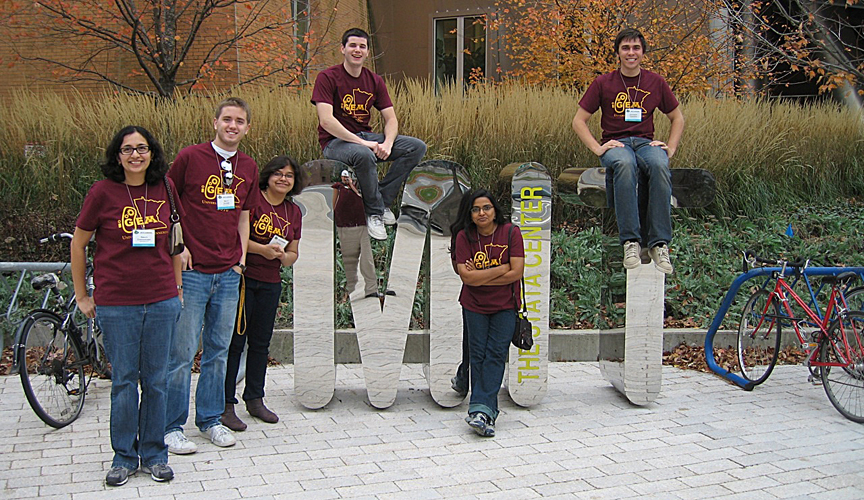|
|
| (38 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| - | <!-- *** What falls between these lines is the Alert Box! You can remove it from your pages once you have read and understood the alert *** -->
| + | [[Image:mnlogo.jpg|300px|center|frame]] |
| | | | |
| - | <html>
| + | {| style="color:gold;background-color:#800000;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="90%" align="center" |
| - | <div id="box" style="width: 700px; margin-left: 137px; padding: 5px; border: 3px solid #000; background-color: #fe2b33;">
| + | !align="center"|[[Team:Minnesota|<font color="gold">Home</font>]] |
| - | <div id="template" style="text-align: center; font-weight: bold; font-size: large; color: #f6f6f6; padding: 5px;">
| + | !align="center"|[[Team:Minnesota/Team|<font color="gold">Team</font>]] |
| - | This is a template page. READ THESE INSTRUCTIONS.
| + | !align="center"|[[Team:Minnesota/Project|<font color="gold">Project</font>]] |
| - | </div> | + | !align="center"|[[Team:Minnesota/Protocols|<font color="gold">Protocols</font>]] |
| - | <div id="instructions" style="text-align: center; font-weight: normal; font-size: small; color: #f6f6f6; padding: 5px;">
| + | !align="center"|[[Team:Minnesota/Notebook|<font color="gold">Notebook</font>]] |
| - | You are provided with this team page template with which to start the iGEM season. You may choose to personalize it to fit your team but keep the same "look." Or you may choose to take your team wiki to a different level and design your own wiki. You can find some examples <a href="https://2009.igem.org/Help:Template/Examples">HERE</a>.
| + | !align="center"|[[Team:Minnesota/Judging|<font color="gold">Judging Criteria</font>]] |
| - | </div> | + | !align="center"|[[Team:Minnesota/Attributions|<font color="gold">Attributions</font>]] |
| - | <div id="warning" style="text-align: center; font-weight: bold; font-size: small; color: #f6f6f6; padding: 5px;">
| + | !align="center"|[[Team:Minnesota/Safety|<font color="gold">Safety</font>]] |
| - | You <strong>MUST</strong> have a team description page, a project abstract, a complete project description, a lab notebook, and a safety page. PLEASE keep all of your pages within your teams namespace.
| + | |} |
| - | </div>
| + | {|align="justify" |
| - | </div> | + | <h1>Welcome!</h1> |
| - | </html> | + | Welcome to the Team Minnesota Wiki for iGEM 2010! |
| | | | |
| - | <!-- *** End of the alert box *** -->
| + | We are a team of undergraduate students along with many faculty and post-doc advisors. This is the third year that Minnesota has sent a team to iGEM. |
| | | | |
| | + | [[Image:MN Igem10 team.jpg|center]] |
| | | | |
| | + | Team Minnesota take won a gold medal and the Best Natural Part Special Prize at iGEM 2010!! |
| | | | |
| - | {|align="justify"
| |
| - | |You can write a background of your team here. <h1>Welcome!</h1>
| |
| - | Welcome to the Team Minnesota Wiki for iGEM 2009!
| |
| | | | |
| - | We are a team of undergraduate and graduate students along with many advisors. This is the second year that Minnesota has sent a team to iGEM. Last year we were runners-up for the Best New BioBrick part (Natural) and we hope to accomplish even more with our design of a logical AND gate in <i>E. coli</i> this year.
| + | <b>Metabolic Engineering: In vivo Nanobioreactors</b> |
| | | | |
| - | <h2>The Mission</h2>
| + | Modern microbial engineering methods allow the introduction of useful exogenous metabolic pathways into cells. Metabolism of certain organic compounds is sometimes limited by the production of toxic intermediates. Several bacteria have evolved protein based microcompartments capable of sequestering such reactions, thus protecting cytosolic machinery and processes from interference by these intermediates. For our iGEM project, we have cloned (and expressed in E. coli) Salmonella LT2 genes responsible for the production and assembly of ethanolamine utilization microcompartments. Additionally, we have determined a signal sequence that targets an ethanolamine utilization enzyme to the microcompartment and verified this by fusing the sequence to GFP and observing that this causes the GFP reporter to localize to the compartment. We conclude that recombinant microcompartments housing targeted enzymes can function as in vivo bioreactors with high reaction efficiencies. |
| - | <p>We are working on a very well studied AND gate, which is composed of elements of the Tet, Lac, and lambda-phage promoters and is responsive to the commonly-used inducers IPTG and aTc, producing GFP as an output signal.
| + | |
| - | Well, we have decided not to ride the wave of ever-more complex synthetic constructs. Instead, we will isolate elements of the AND gate and study them individually. This will allow for a greater understanding of how exactly this AND gate works, and will eventually allow for creation of better AND gates, and a more predictable response to inputs by the biological systems.</p>
| + | |
| - | <p>Our group will be doing both experimental and modeling work this year. These two approachs will complement each other. The experimental data will be used to refine the mathematical models, while the models will determine good directions for the experiments to pursue. We will go into greater detail on both these aspects on theeir respective wiki pages. The approach we are taking can be applied to many other biological circuits and can lead to greater understanding of how to engineer biological circuits.</p>
| + | |
| - | <p> What is unique in our work is the effort to develop software tools that streamline the construction of synthetic biological systems. The Synthetic Biology Software Suite is the product of our work, and this year iGEM students are working on SynBioSS Designer, a tool that uses Registry parts to build a model of biomolecular interaction. This model can be used to simulate the dynamic behavior of the gene network.</p>
| + | |
| | | | |
| - | <h2>Highlights for the Judges</h2>
| |
| - | Judges! Please navigate to our [https://2009.igem.org/Team:Minnesota/Criteria Judging Criteria] page for a comprehensive list of UMN's fulfilled medal requirements and links throughout the wiki.
| |
| | | | |
| - | |[[Image:Minnesota_logo.png|200px|right|frame]]
| |
| - | |-
| |
| - | |
| |
| - | ''Tell us more about your project. Give us background. Use this as the abstract of your project. Be descriptive but concise (1-2 paragraphs)''
| |
| - | |[[Image:Minnesota_team.png|right|frame|Your team picture]]
| |
| - | |-
| |
| - | |
| |
| - | |align="center"|[[Team:Minnesota | Team Example]]
| |
| - | |}
| |
| | | | |
| - | <!--- The Mission, Experiments ---> | + | <h2>Highlights for the Judges</h2> |
| | + | Judges! Please navigate to our [[Team:Minnesota/Judging|Judging Criteria]] page for a comprehensive list of UMN's fulfilled medal requirements and links throughout the wiki. |
| | | | |
| - | {| style="color:gold;background-color:#800000;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="90%" align="center" | + | {|align="center" cellpadding="25" |
| - | !align="center"|[[Team:Minnesota|<font color="gold">Home</font>]]
| + | |[[Image:UMNSponsors.png|300px|center|frame|Sponsers]] |
| - | !align="center"|[[Team:Minnesota/Team|<font color="gold">The Team</font>]]
| + | |
| - | !align="center"|[[Team:Minnesota/Project|<font color="gold">The Project</font>]]
| + | |
| - | !align="center"|[[Team:Minnesota/Designer|<font color="gold">SynBioSS Designer</font>]]
| + | |
| - | !align="center"|[[Team:Minnesota/Modeling|<font color="gold">Modeling</font>]]
| + | |
| - | !align="center"|[[Team:Minnesota/Notebook|<font color="gold">Experimental</font>]]
| + | |
| - | !align="center"|[[Team:Minnesota/Parts Characterization|<font color="gold">Competition Requirements</font>]]
| + | |
| - | |}
| + | |
Welcome!
Welcome to the Team Minnesota Wiki for iGEM 2010!
We are a team of undergraduate students along with many faculty and post-doc advisors. This is the third year that Minnesota has sent a team to iGEM.
Team Minnesota take won a gold medal and the Best Natural Part Special Prize at iGEM 2010!!
Metabolic Engineering: In vivo Nanobioreactors
Modern microbial engineering methods allow the introduction of useful exogenous metabolic pathways into cells. Metabolism of certain organic compounds is sometimes limited by the production of toxic intermediates. Several bacteria have evolved protein based microcompartments capable of sequestering such reactions, thus protecting cytosolic machinery and processes from interference by these intermediates. For our iGEM project, we have cloned (and expressed in E. coli) Salmonella LT2 genes responsible for the production and assembly of ethanolamine utilization microcompartments. Additionally, we have determined a signal sequence that targets an ethanolamine utilization enzyme to the microcompartment and verified this by fusing the sequence to GFP and observing that this causes the GFP reporter to localize to the compartment. We conclude that recombinant microcompartments housing targeted enzymes can function as in vivo bioreactors with high reaction efficiencies.
Highlights for the Judges
Judges! Please navigate to our Judging Criteria page for a comprehensive list of UMN's fulfilled medal requirements and links throughout the wiki.
 Sponsers |
|
 "
"