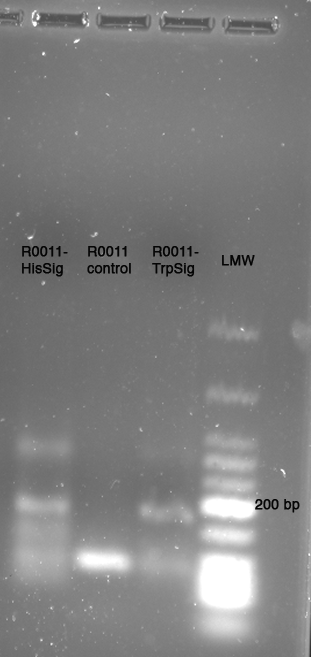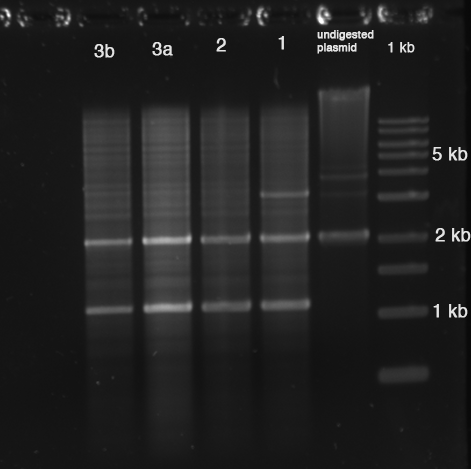Team:TU Munich/Lab
From 2010.igem.org
FPraetorius (Talk | contribs) (→24.09.2010) |
FPraetorius (Talk | contribs) (→Lab Book) |
||
| (83 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
<!-- ############## WIKI-PAGE STARTS HERE ############## --> | <!-- ############## WIKI-PAGE STARTS HERE ############## --> | ||
=Experiment Design= | =Experiment Design= | ||
| - | In this section we do not only want to present the experiments and results we gained but also to encourage you to evaluate your own | + | In this section we do not only want to present the experiments and results we gained but also to encourage you to evaluate your own switch based on the protocols and general procedure on how to evaluate basic parameters of a switch. In theory, every terminator can be turned into a switch with minor modifications and the right signals which are based on individual applications. While the principle of how to turn a terminator into a switch is explained in detail [https://2010.igem.org/Team:TU_Munich/Project here], experimental setups and protocols are provided in the following. Due to time and equipment limitations we could not perform all the experiments we planned but next to the hope that another iGEM team might proceed with our project we would also like to encourage you to design and test some basic switches on which you can base a complete, tightly regulated network. |
| - | + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | |
| - | The complexity of our experimental setups vary, since we planned to characterize an individual | + | The complexity of our experimental setups vary, since we planned to characterize an individual switch with one exemplary signal on all relevant levels: Starting from the most general, complicated but also relevant level, ''in vivo'' measurements we approached to testing different switches on each smaller scale: We developed setups for ''in vitro'' translation which can be done without much effort following the ''in vitro'' measurements and also provide detailed description of ''in vitro'' transcription verification providing an inside to the molecular functionality of our basic idea. We do not see the methods we used here as the gold standard for bioLOGICS evaluation and encourage you to include your own ideas as well as check in our outlook section where we suggest experiments we could not do during the limited iGEM 2010 time. Together with our [https://2010.igem.org/Team:TU_Munich/Parts Biobrick submissions] this year, we offer a complete set for switch evaluation on all cellular levels. |
<br> | <br> | ||
Most measurements are based on fluorescence reporters which provide easy handling, fast output and are well studied. Next to the fluorescent proteins GFP and mCherry we used ''in vivo'', a malachite green binding aptamer serves as a reporter ''in vitro'' providing a reliable fluorescent output upon antitermination. | Most measurements are based on fluorescence reporters which provide easy handling, fast output and are well studied. Next to the fluorescent proteins GFP and mCherry we used ''in vivo'', a malachite green binding aptamer serves as a reporter ''in vitro'' providing a reliable fluorescent output upon antitermination. | ||
<br> | <br> | ||
| - | Most setups up to now were only used to evaluate | + | Most setups up to now were only used to evaluate switches with an default state "off" which are applied for AND/OR devices. In principle the same methods can be used for NOT devices which are based on a switch with an default state "off". Again, time limitation circumvented further tested from our team but we hope that further studies can be done in the future. |
<!-- Our initial idea to prove our concept of antitermination was to use fluorescent proteins as reporters. This approach gives the opportunity to measure the termination and antitermination efficiency of our designed BioBricks ''in vivo'' as well as ''in vitro'', the latter using a translation kit based on <i>E. coli</i> lysate. Later on, we decided to develop an experiment, that relies only on transcription. In this set-up, we used a fluorescent dye, malachite green, that binds a specific RNA aptamer and thus makes it possible to detect transcription activity. --> | <!-- Our initial idea to prove our concept of antitermination was to use fluorescent proteins as reporters. This approach gives the opportunity to measure the termination and antitermination efficiency of our designed BioBricks ''in vivo'' as well as ''in vitro'', the latter using a translation kit based on <i>E. coli</i> lysate. Later on, we decided to develop an experiment, that relies only on transcription. In this set-up, we used a fluorescent dye, malachite green, that binds a specific RNA aptamer and thus makes it possible to detect transcription activity. --> | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
==''In vivo'' Measurements== | ==''In vivo'' Measurements== | ||
| Line 21: | Line 22: | ||
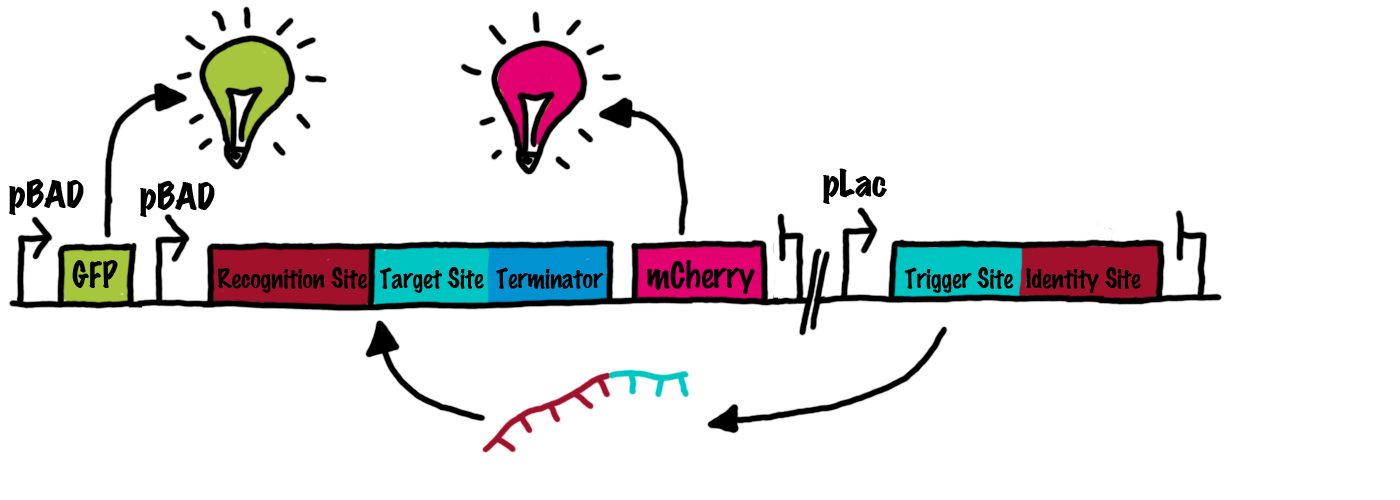
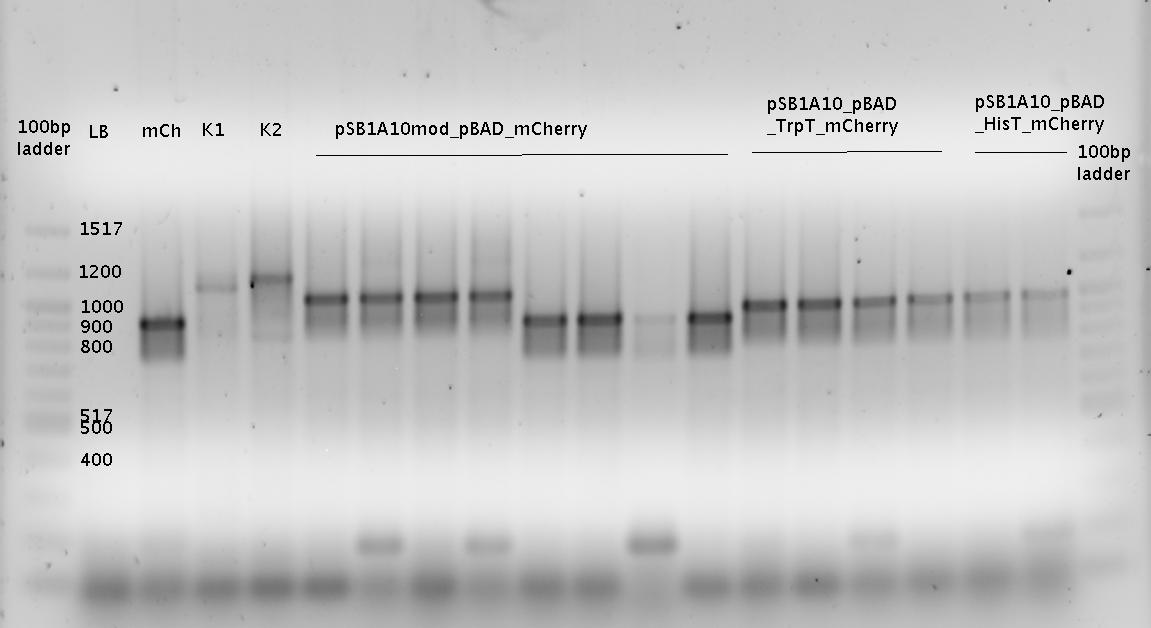
For the measurements ''in vivo'' we decided to use an expression cassette consisting of Green Fluorescent Protein (GFP) coding sequence upstream of the switch and another fluorescent protein coding sequence downstream of it. Both protein coding sequence share the same ribosome binding site allowing the usage of the GFP as an internal control in measurements. Since the spectra should not overlap and to avoid FRET as well as an pure overlap of the spectra, we settled on the usage of red fluorescent protein variants, namely mRFP1 in the first try and mCherry in an modified variant of the pSB1A10 vector. | For the measurements ''in vivo'' we decided to use an expression cassette consisting of Green Fluorescent Protein (GFP) coding sequence upstream of the switch and another fluorescent protein coding sequence downstream of it. Both protein coding sequence share the same ribosome binding site allowing the usage of the GFP as an internal control in measurements. Since the spectra should not overlap and to avoid FRET as well as an pure overlap of the spectra, we settled on the usage of red fluorescent protein variants, namely mRFP1 in the first try and mCherry in an modified variant of the pSB1A10 vector. | ||
<br> | <br> | ||
| - | While the GFP fluorescence can be used to normalize the measurements, the RFP fluorescence serves as a reporter to detect and evaluate termination and antitermination. To stimulate the expression of the fluorescent proteins, we took advantage of the pBAD promoter family (sensitive towards arabinose). The signal upon which the antitermination events and therefore switching relies | + | While the GFP fluorescence can be used to normalize the measurements, the RFP fluorescence serves as a reporter to detect and evaluate termination and antitermination. To stimulate the expression of the fluorescent proteins, we took advantage of the pBAD promoter family (sensitive towards arabinose). The signal upon which the antitermination events and therefore switching relies on were under the control of an IPTG inducible promoter. We went with this well-established pair of controllable promoters to deliver an easy setup in the beginning, like described [https://2010.igem.org/Team:TU_Munich/Software here], every sort of input may later be combined with our basic switching units. |
<br> | <br> | ||
| - | [[Image: | + | [[Image:invivo3.png|500px|thumb|center|general measurement principle]] |
<br> | <br> | ||
The GFP internal control carries the advantage that errors in the measurement set can be detected easily. Lack of arabinose or promoter insensitivity can be recognized as well as problems with the fluorescence measurement itself. Plus, it allows normalizing measurements to compare different preparations in relation to each other. | The GFP internal control carries the advantage that errors in the measurement set can be detected easily. Lack of arabinose or promoter insensitivity can be recognized as well as problems with the fluorescence measurement itself. Plus, it allows normalizing measurements to compare different preparations in relation to each other. | ||
<br> | <br> | ||
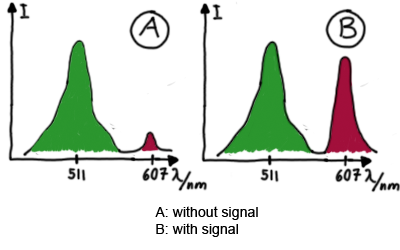
| - | Upon binding of a signal to the terminator | + | Upon binding of a signal to the terminator switch, termination is circumvented and the reporter protein behind the switch can be translated. In the experimental setup presented here, this will result in an RFP expression, but again, every protein or DNA-encoded element in general may be used as an output. Since the RFP fluorescence spectra do not overlap with GFP it offers an easy possibility to evaluate the effect of signal induction. Next to GFP fluorescence, RFP fluorescence will show up. |
| - | [[Image:TUM2010_Expected_emission_spextra.png|center| | + | <br> |
| + | [[Image:TUM2010_Expected_emission_spextra.png|375px|thumb|center|schematic estimated fluorescence spectra]] | ||
<br> | <br> | ||
===Construction and Cloning=== | ===Construction and Cloning=== | ||
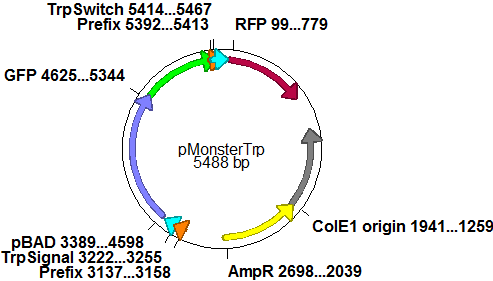
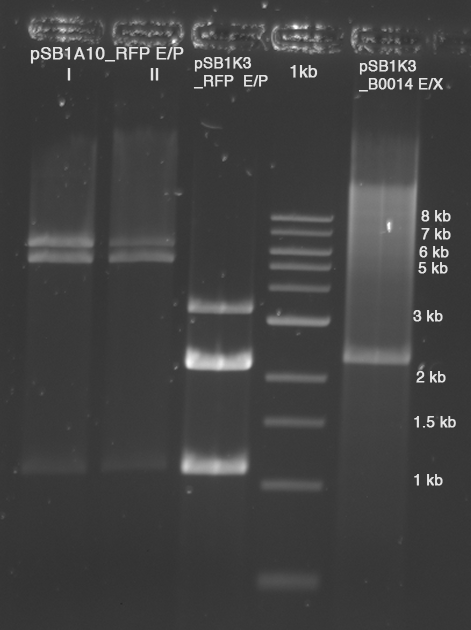
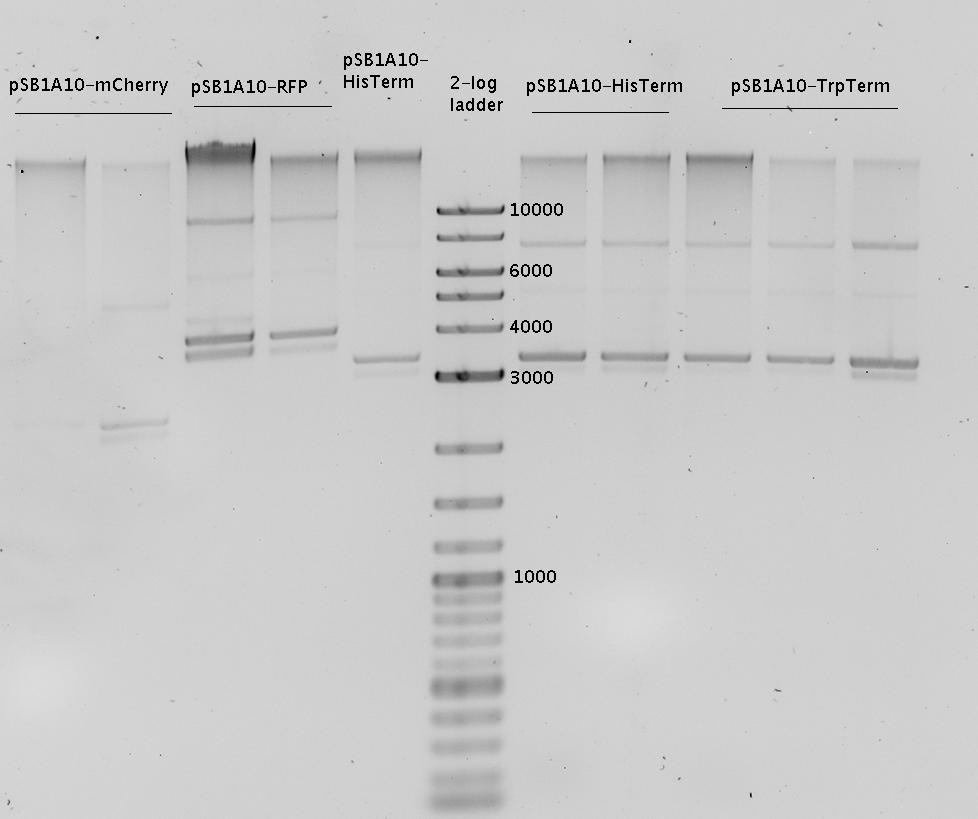
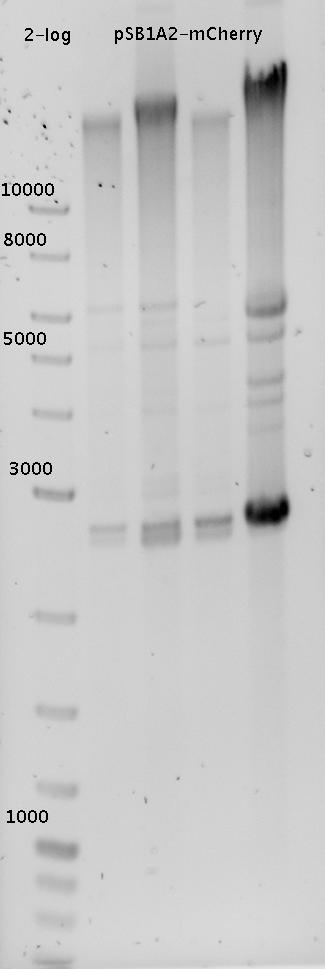
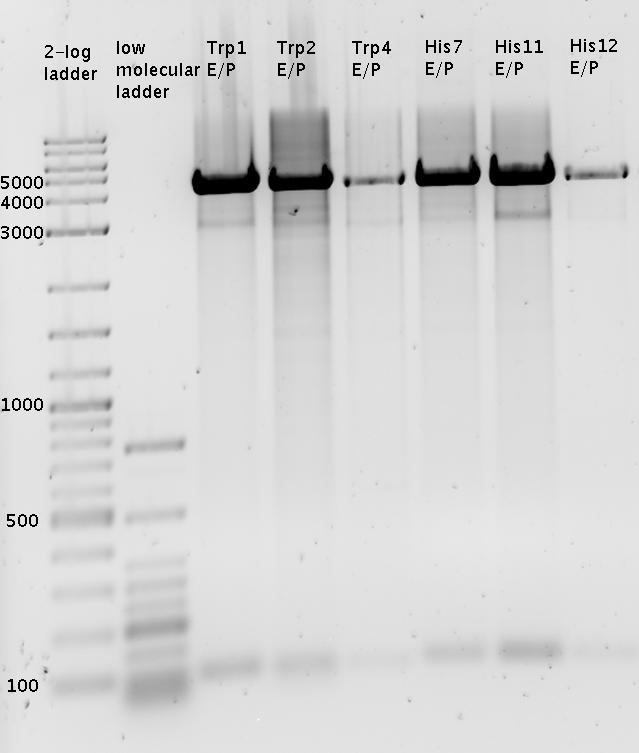
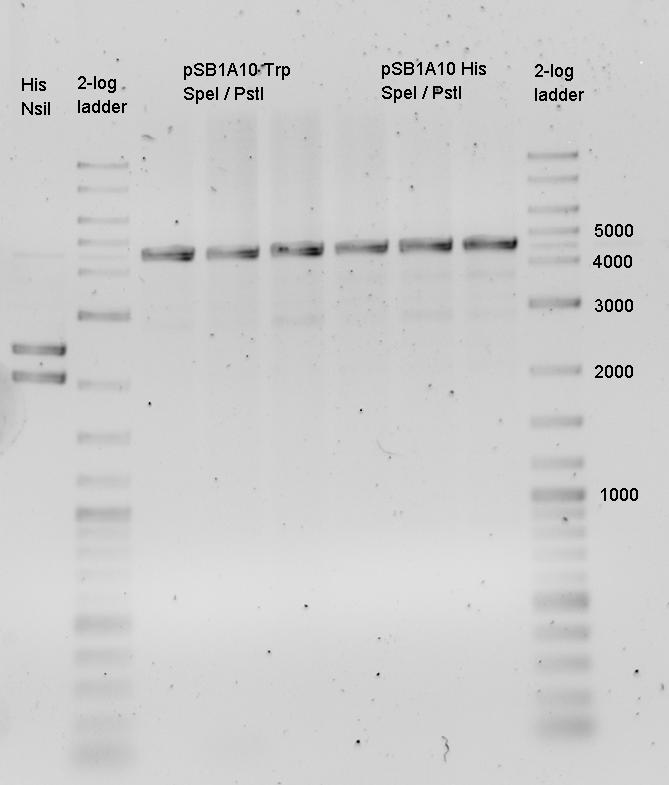
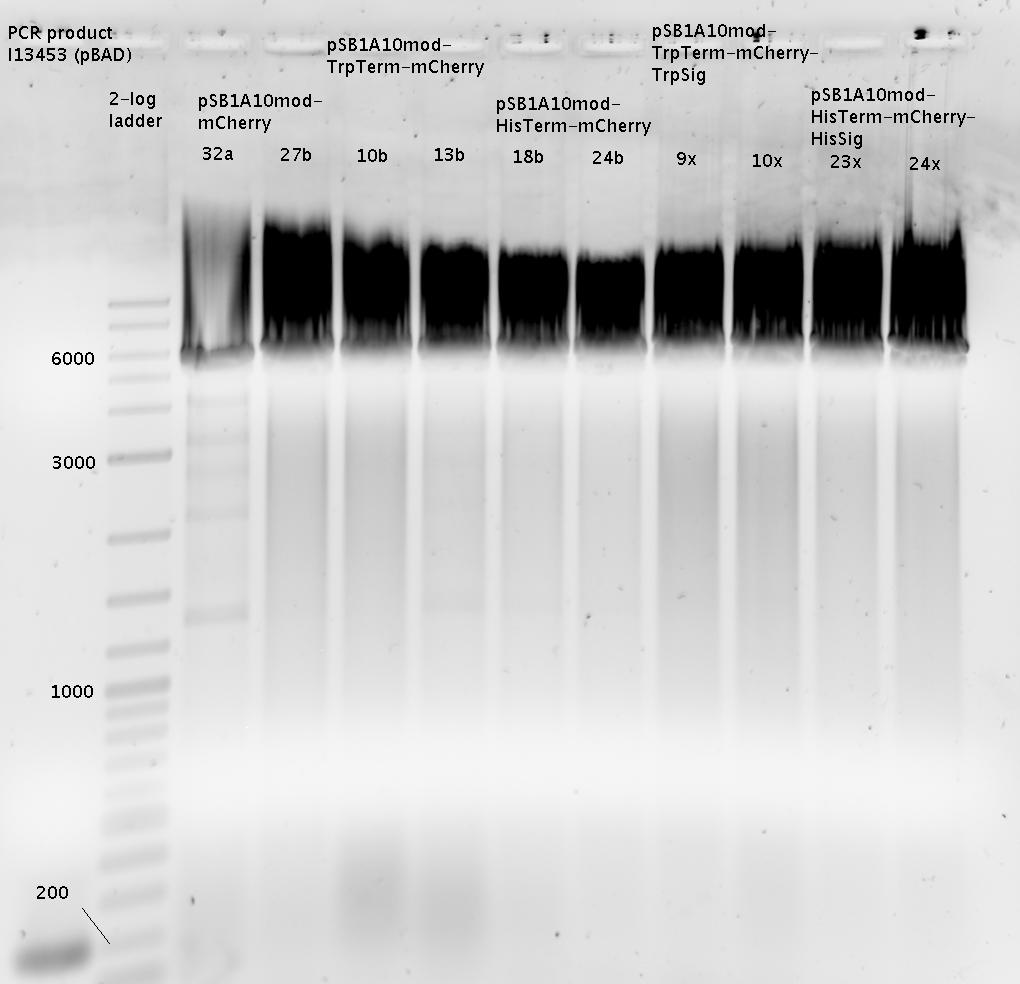
| - | In a first try we cloned a measuring | + | In a first try we cloned a measuring construct based on pSB1A10. The resulting plasmid, nicknamed pMonsterplasmid due to its size was tested in the fluorescent measurements described [https://2010.igem.org/Team:TU_Munich/Lab#Switch_evaluation_in_vivo below]. Unfortunately after two months of cloning we had to recognize that the plasmid in use did not work for us (see also [[Team:TU_Munich/Parts#Falsified_Parts|pSB1A10 Falsification]]). <br> |
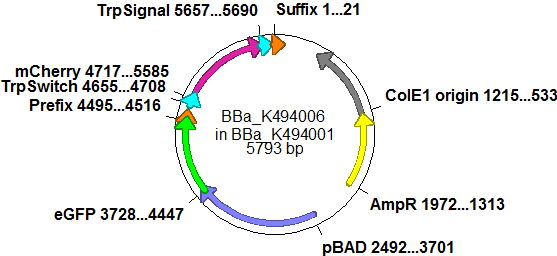
| - | So after the first unsuccessful attempts we decided to reclone the system, | + | So after the first unsuccessful attempts we decided to reclone the system, substituting RFP to mCherry, a dsRED derivative with a spectrum in the far red, and adding arabinose inducible promoters in front of both fluorescent proteins to guarantee stable and comparable expression of both proteins |
{| | {| | ||
| - | |[[Image:TUM2010 | + | |[[Image:TUM2010 Plasmid1flo.png|350px|right|first measuring construct]] |
| - | | | + | |[[Image:TUM2010 Plasmid2flo.png|350px|left|BBa_K494006 cloned in BBa_K494001]] |
|} | |} | ||
<br> | <br> | ||
| - | To control the expression of the switch, the particular DNA sequence itself is under the control of an IPTG dependent promoter. In the future we want our networks to be able to respond to a variety of external signals like small metabolites, ions or whatever can be found in the parts registry. For basic | + | To control the expression of the switch, the particular DNA sequence itself is under the control of an IPTG dependent promoter. In the future we want our networks to be able to respond to a variety of external signals like small metabolites, ions or whatever can be found in the parts registry. For basic switch evaluation, an established and well-working system like the ''lac''-operon was chosen to avoid side-effects of less well-characterized promoters. |
<br> | <br> | ||
===Measurements based on submitted Biobricks=== | ===Measurements based on submitted Biobricks=== | ||
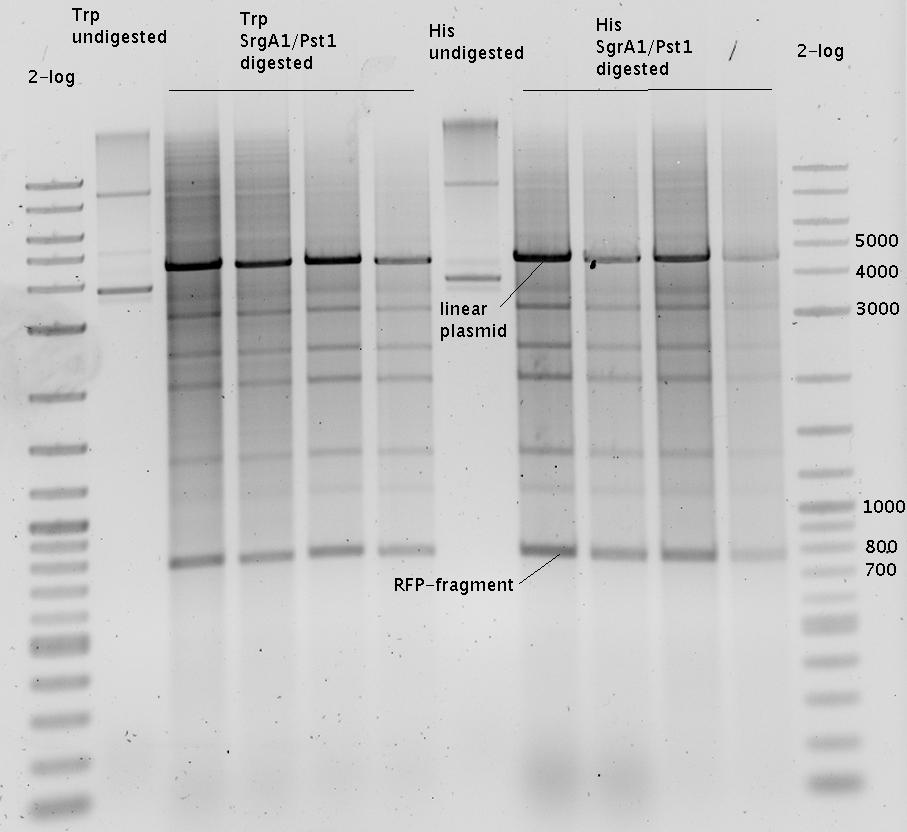
| - | The Biobricks BBa_K494001-BBa_K494006 are constructed for easy design of | + | The Biobricks BBa_K494001-BBa_K494006 are constructed for easy design of a switch-evaluation system. Detailed information can be found [https://2010.igem.org/Team:TU_Munich/Parts#Plasmids here]. |
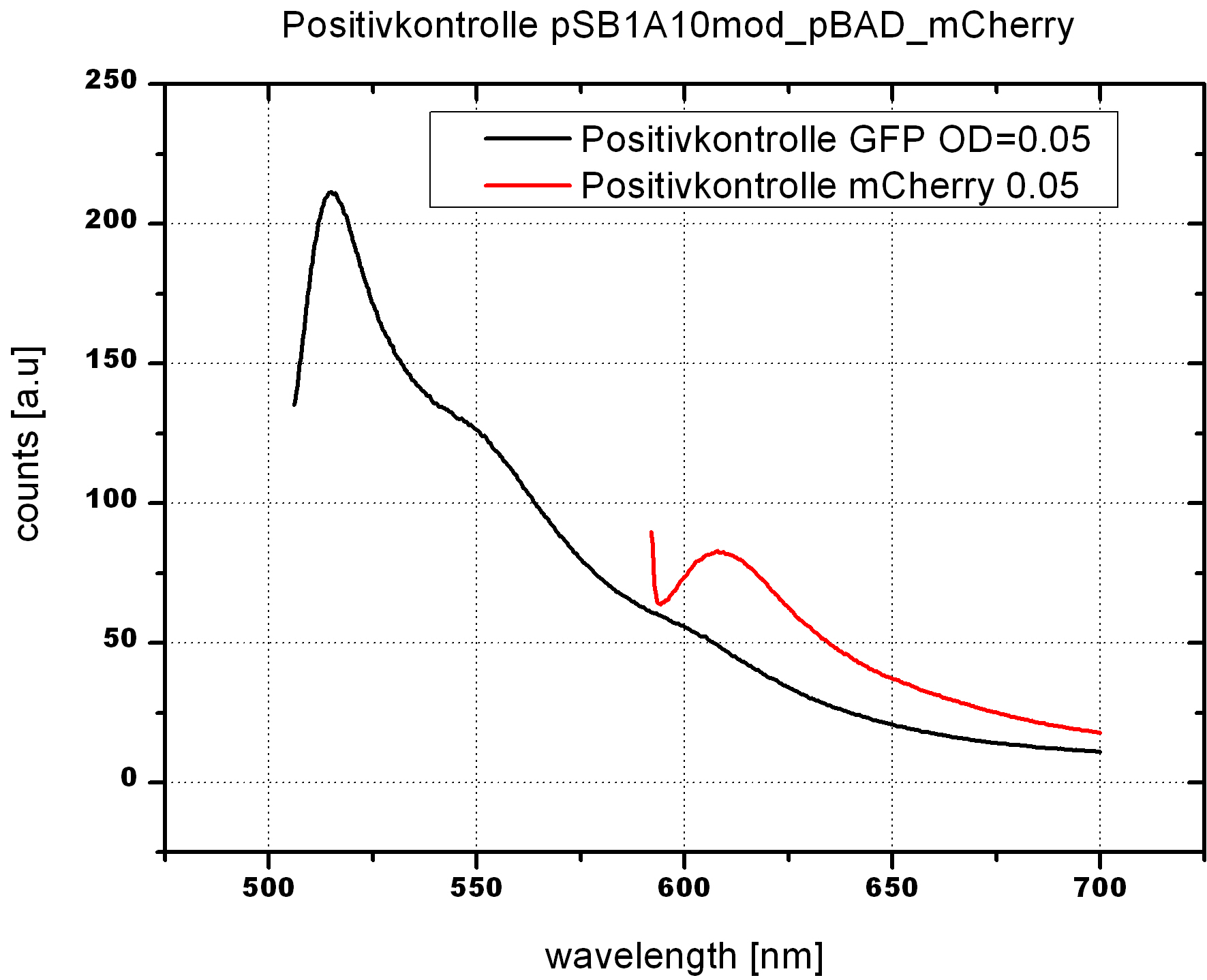
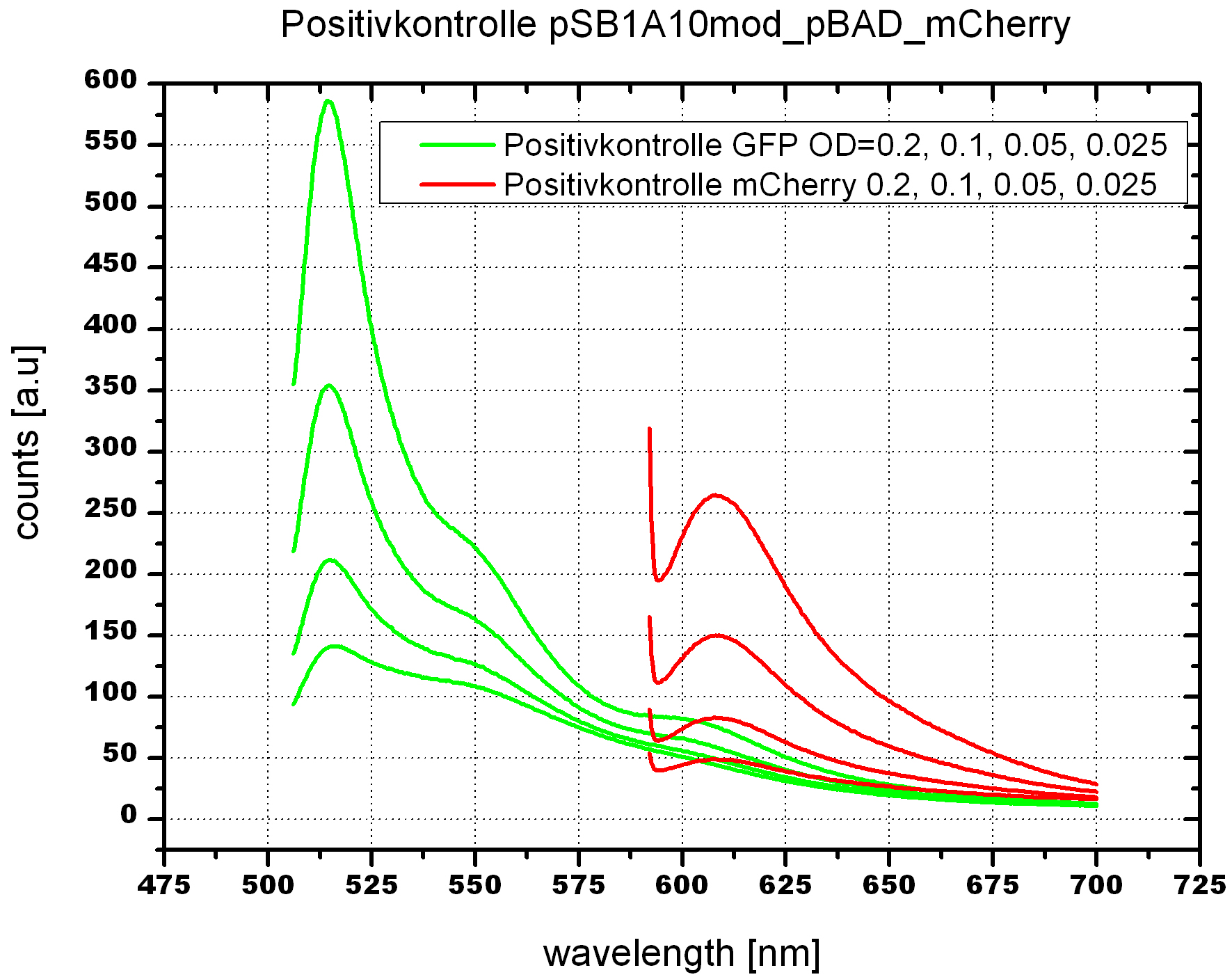
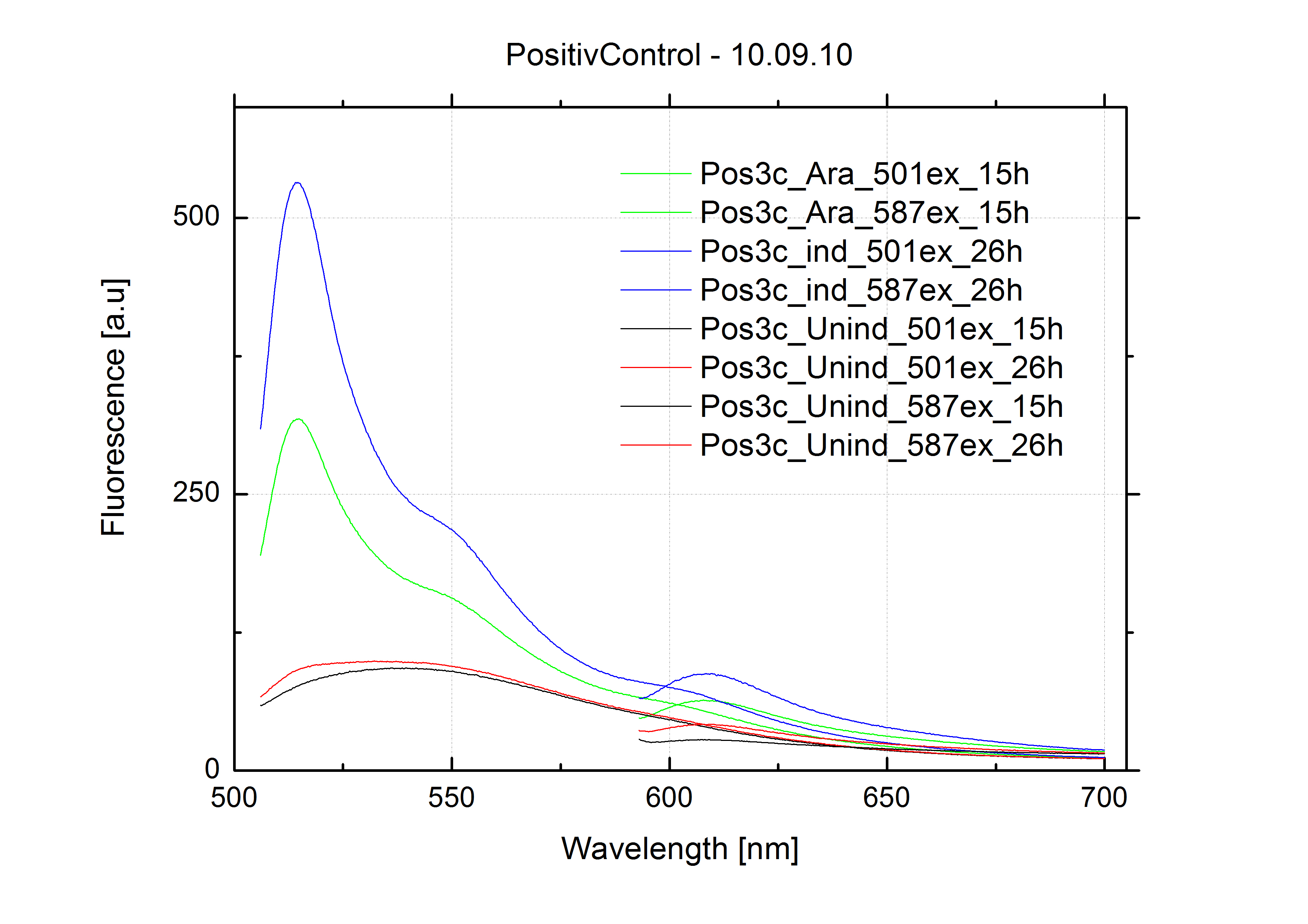
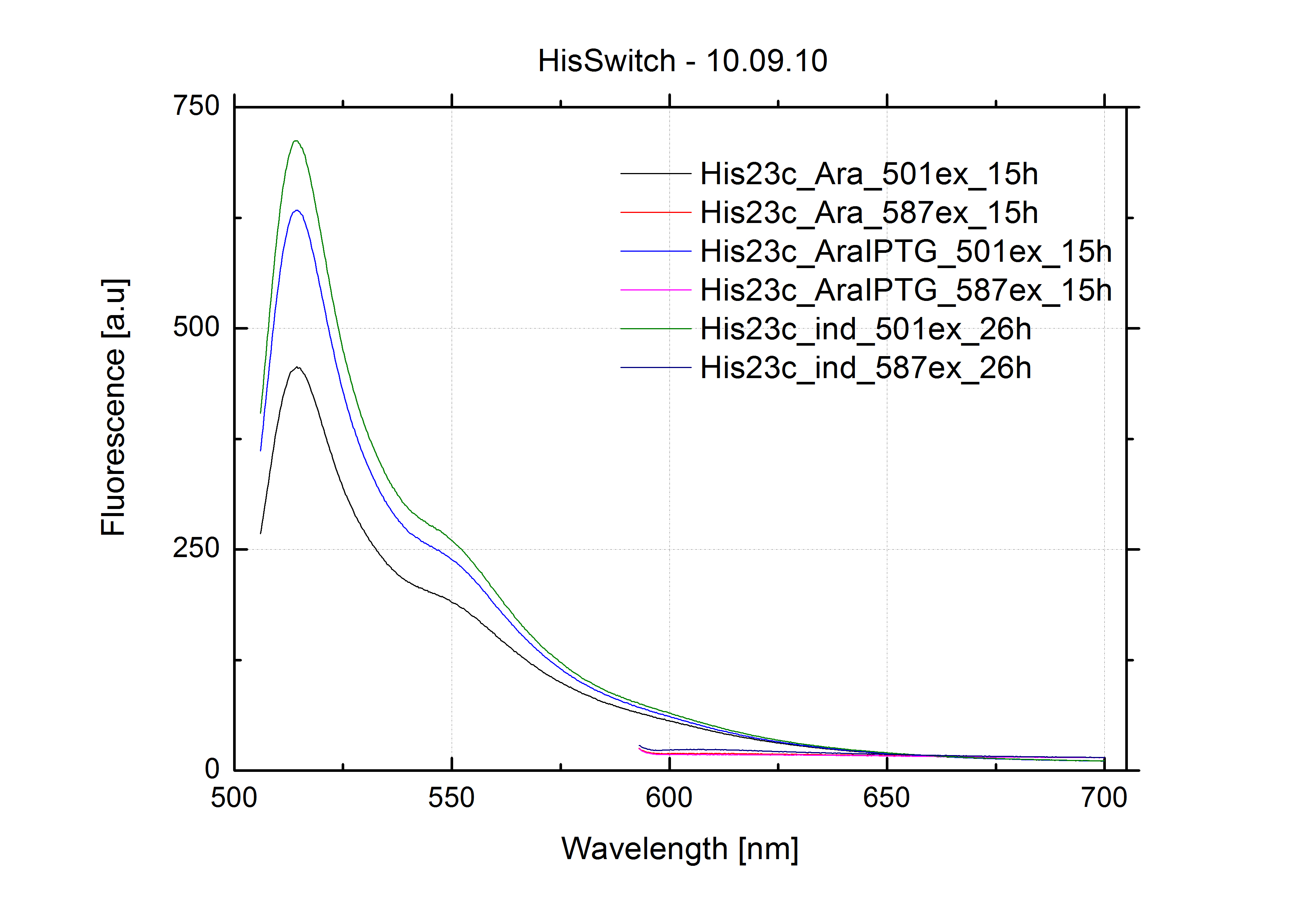
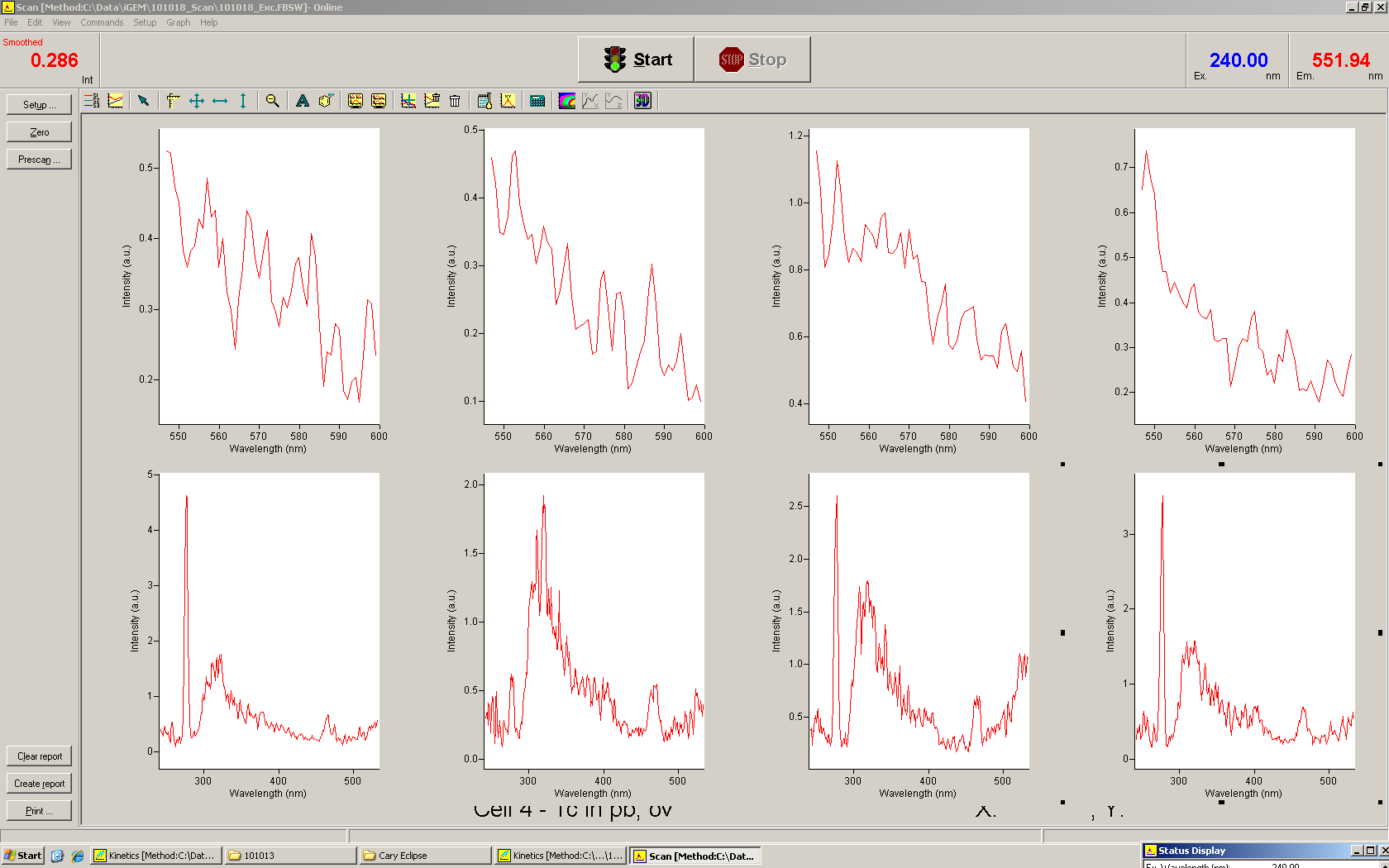
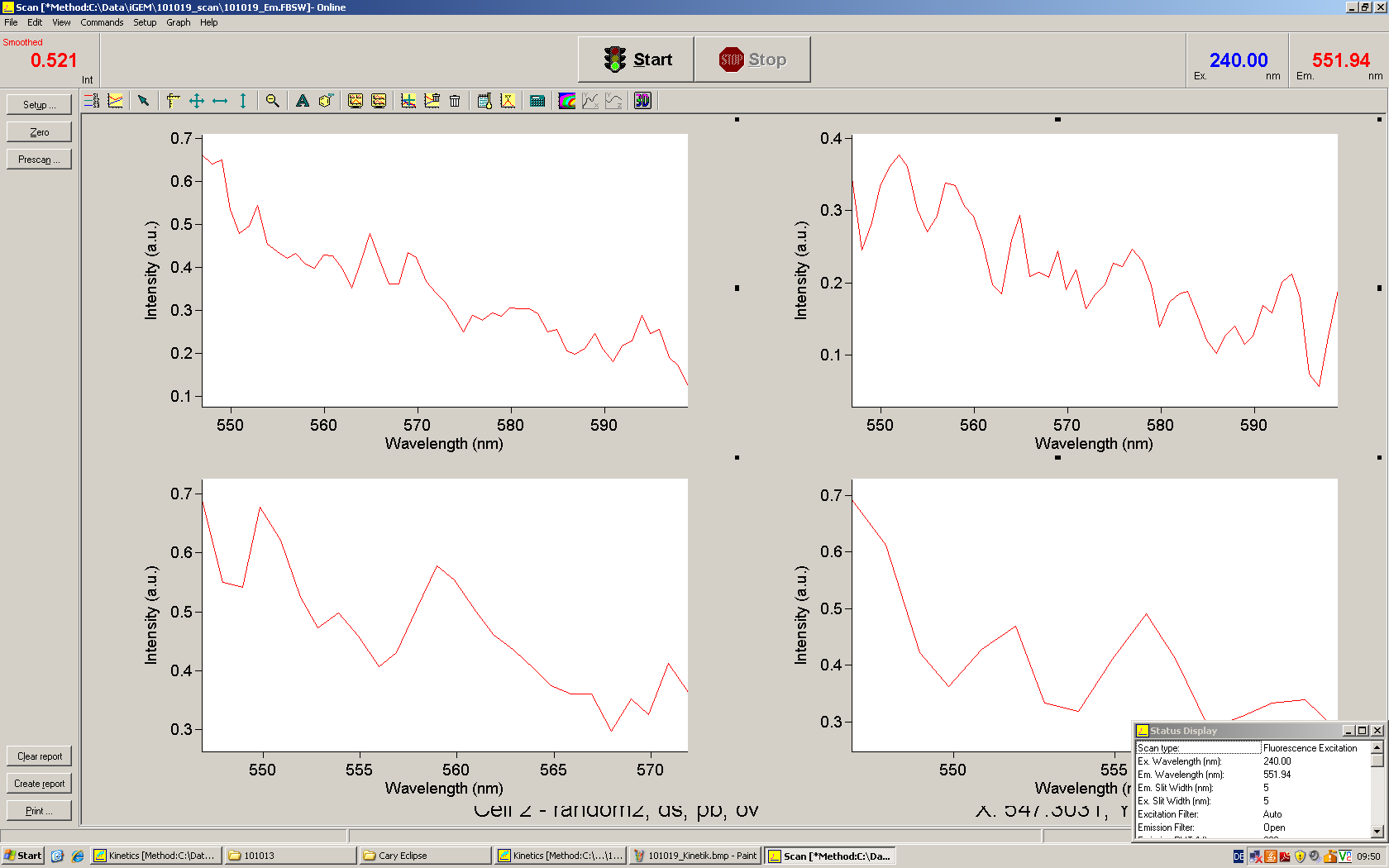
===Switch evaluation ''in vivo''=== | ===Switch evaluation ''in vivo''=== | ||
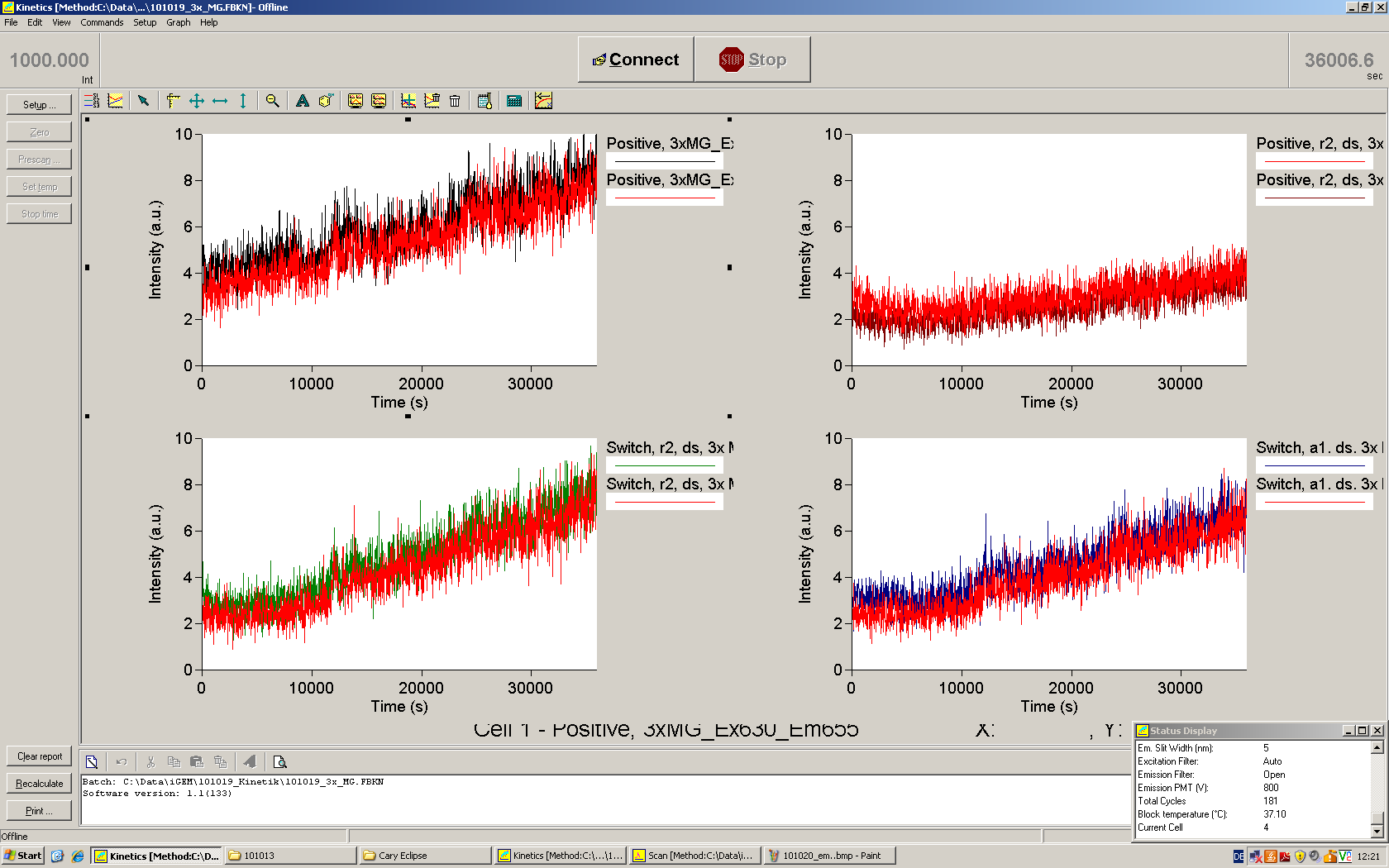
| - | To evaluate the switching efficiency, output with and without signal needs to be monitored. In this case, GFP | + | To evaluate the switching efficiency, output with and without signal needs to be monitored. In this case, GFP fluorescence (internal control) will always appear upon arabinose induction, while RFP/mCherry fluorescence is only present upon binding of a signal and occurring antitermination. <br><br> |
| - | Upon induction with arabinose a rise of GFP expression can be seen. To monitor changes in gene expression we used a fluorimeter and measured fluorescence of whole living cells. While this approach provides easy handling and monitoring, too much scattering has to be carefully avoided: the cell density should not exceed an | + | Upon induction with arabinose a rise of GFP expression can be seen. To monitor changes in gene expression we used a fluorimeter and measured fluorescence of whole living cells. While this approach provides easy handling and monitoring, too much scattering has to be carefully avoided: the cell density should not exceed an OD<sub>600</sub> of 0.05. RFP/mCherry emission should be visible only in case of a working switch or inefficient termination. |
| + | <br><br> | ||
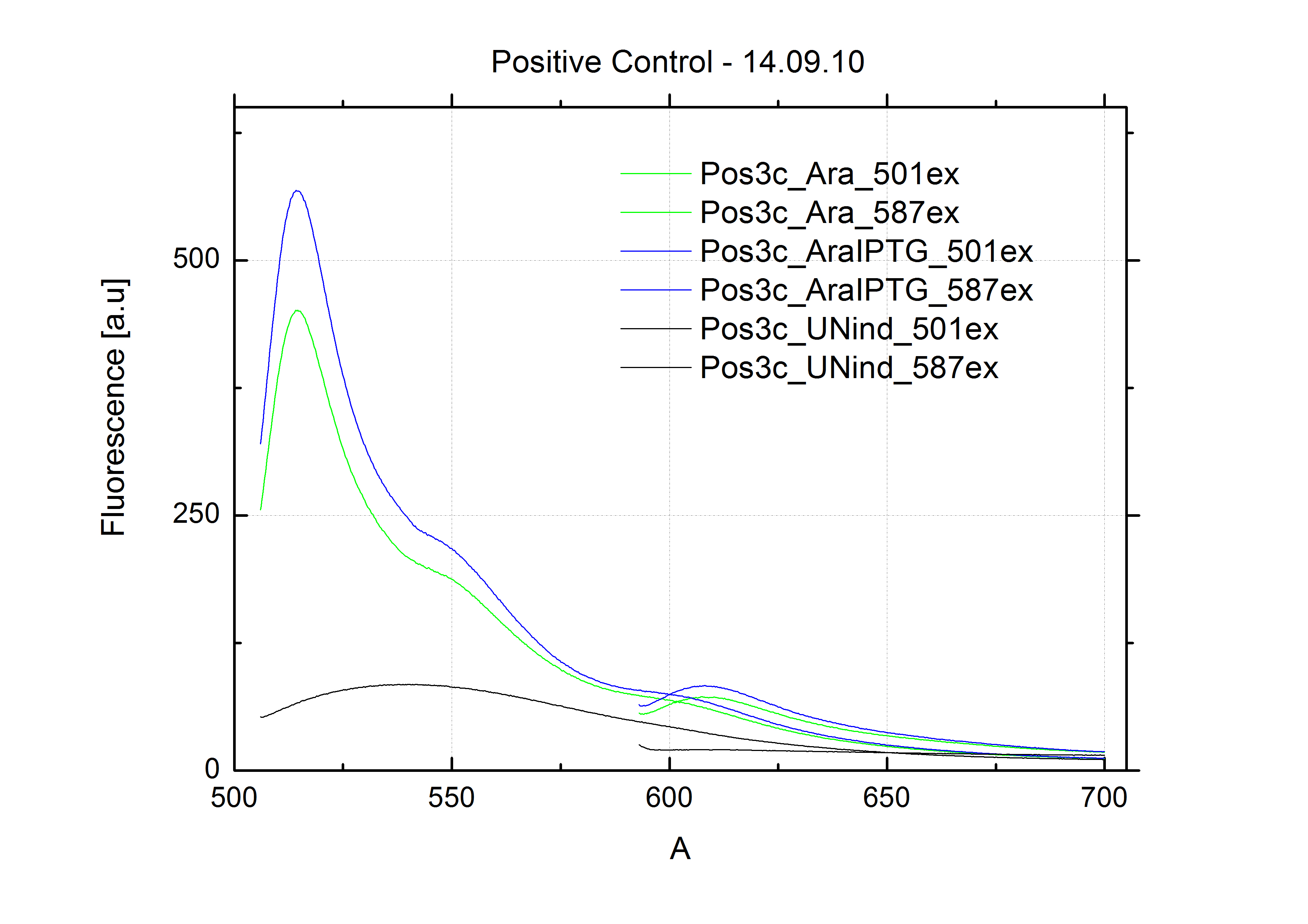
| + | For evaluation of the measuring plasmid itself we incorporated a positive control in every measurement. A random sequence in between GFP and RFP/mCherry was chosen in a corresponding length instead of a terminator. An increase in both GFP and mCherry was detectable in comparable amounts after quantum yield correction, showing the measuring plasmid to beworking nicely. While the positive control may be the same for all evaluated devices, the negative control has to be specific for every switch or terminator, respectively. | ||
<br> | <br> | ||
| - | + | [[image:TUM2010_bacteriaAll.JPG|thumb|375px|center|Bacterial cultures after incubation of 16h]] | |
<br> | <br> | ||
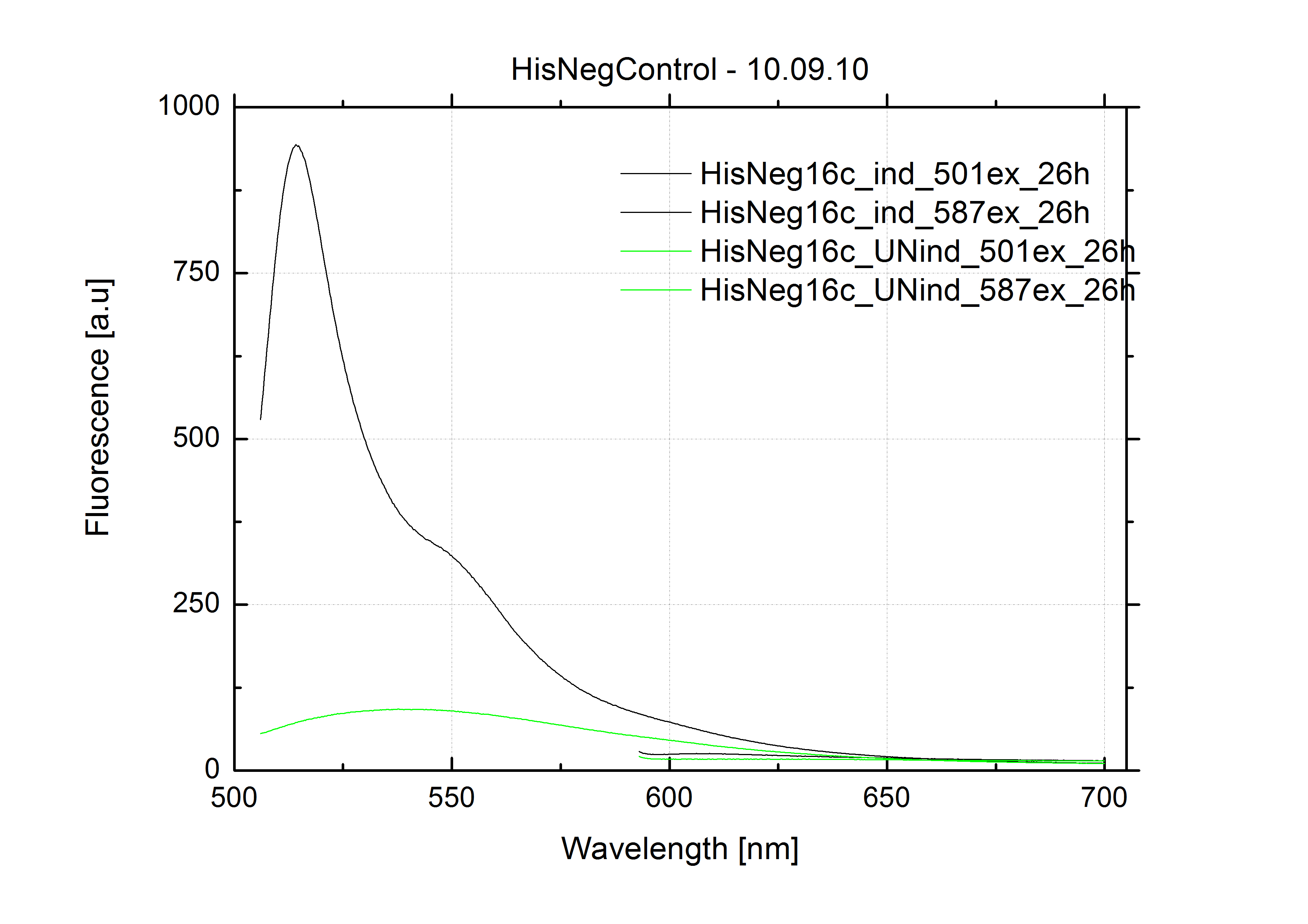
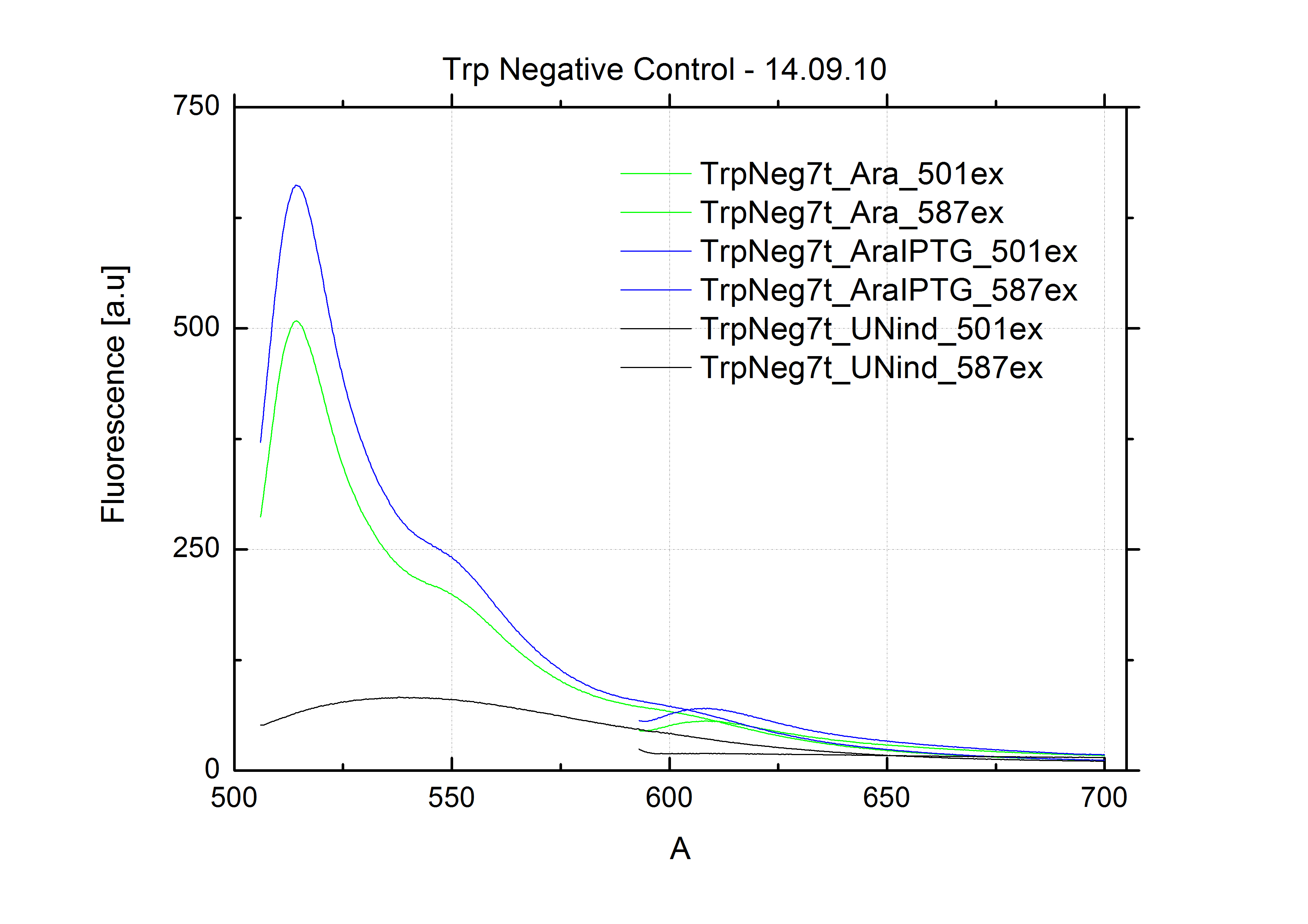
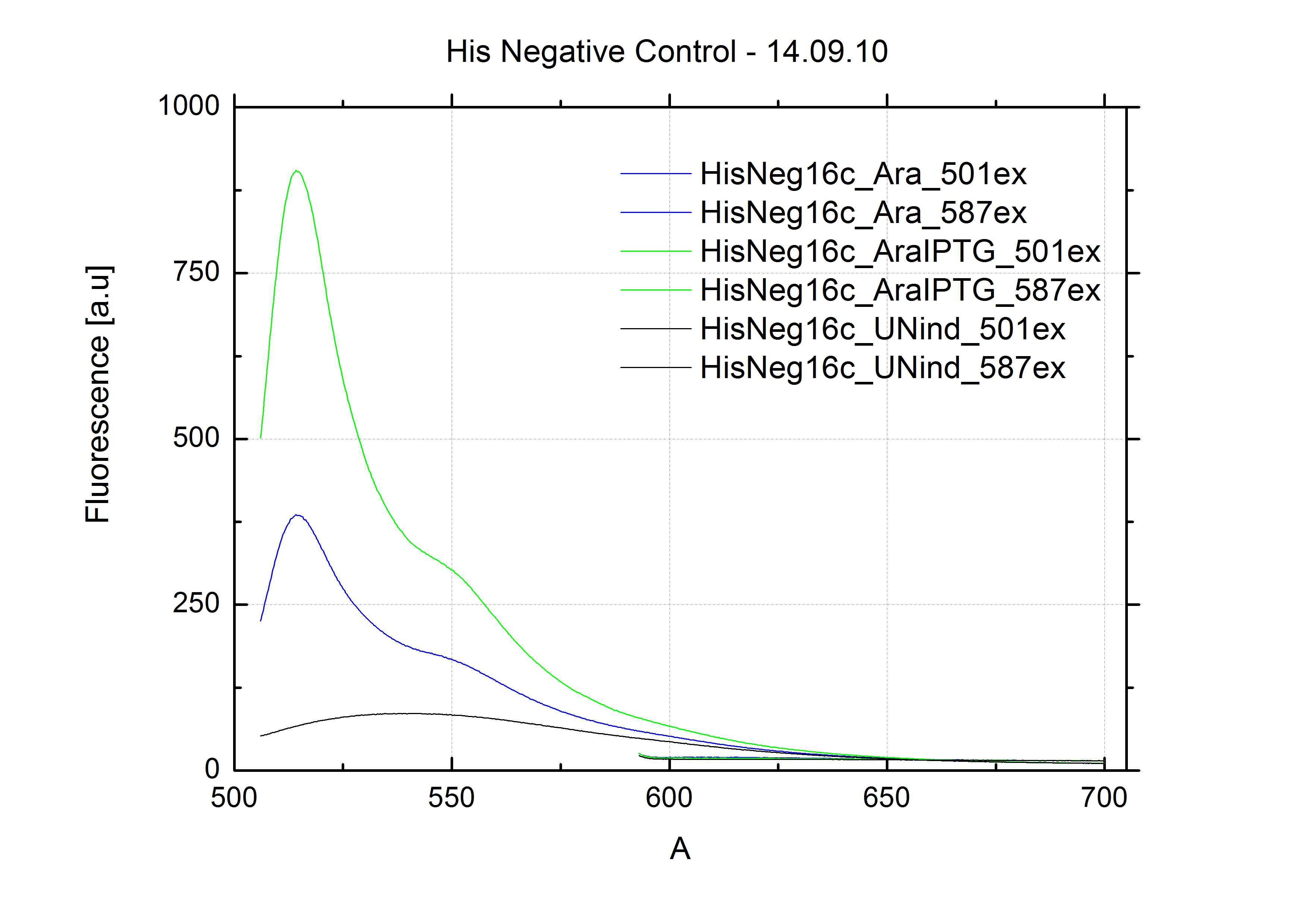
| - | + | The negative control contained the evaluated switch without any possibility for induction of the corresponding signal. Thereby the switch's function is limited to termination, leading to no detectable RFP/mCherry fluorescence. By definition every switch type has to be tested using a negative control without a corresponding signal, since termination efficiency may vary depending on the terminator itself, cell strain and general growth conditions. We recommend to chose your terminator of choice and evaluate it using the provided plasmids. <br> | |
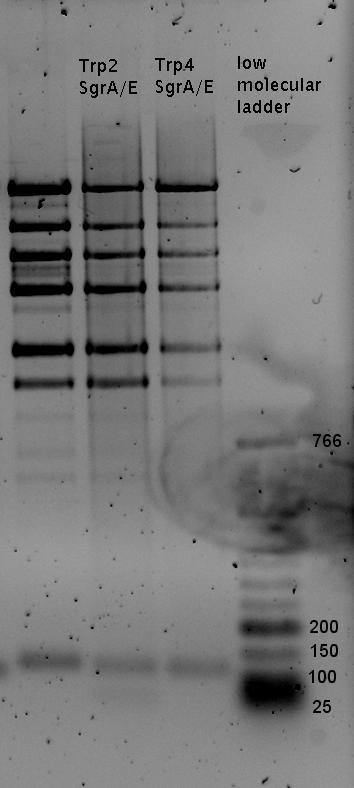
| - | In our experimental part we | + | In our experimental part we evaluated terminators based on the regulatory unit of the tryptophan (Trp-Term) and histidine (His-Term) operons. Those synthetic operons are regulated based on the principle of attenuation, a terminator in front of genes involved in amino acid biosynthesis avoids transcription until environmental stimulis suggest a lack of those amino acids. Since both sequences are known to be regulated by changes in secondary structure, those two attenuators became the basis for our designed switches. <br> |
| + | The terminators we tested can be found in the Parts Registry. With the construction of the backbone [http://partsregistry.org/Part:BBa_K494001 BBa_K494001], potential switches and signals can be easily subcloned in two steps and tested. [http://partsregistry.org/Part:BBa_K494002 BBa_K494002] was constructed as a positive control, producing maximal mCherry fluorescence which may be used to characterize terminator and switch efficiency. [http://partsregistry.org/Part:BBa_K494003 BBa_K494003] and [http://partsregistry.org/Part:BBa_K494004 BBa_K494004] carry the His-Terminator with and without the corresponding signal, [http://partsregistry.org/Part:BBa_K494005 BBa_K494005] and [http://partsregistry.org/Part:BBa_K494006 BBa_K494006] being the same for Trp-Terminator. | ||
| Line 65: | Line 70: | ||
===Design=== | ===Design=== | ||
Since the same constructs can be used both for ''in vivo'' and ''in vitro'' translation, no additional cloning effort is needed. This implements, that the Biobricks we [https://2010.igem.org/Team:TU_Munich/Plasmids provided this year], can again be used as the groundwork for constructing vectors for measurements. <br> | Since the same constructs can be used both for ''in vivo'' and ''in vitro'' translation, no additional cloning effort is needed. This implements, that the Biobricks we [https://2010.igem.org/Team:TU_Munich/Plasmids provided this year], can again be used as the groundwork for constructing vectors for measurements. <br> | ||
| - | Reporter proteins GFP and mCherry are well expressed ''in vitro'', the limiting factors are mostly the capacity of a kit versus the maturation time of fluorescent proteins. Since mCherry was characterized with a maturation time of 15 minutes by Tsien and Coworkers | + | Reporter proteins GFP and mCherry are well expressed ''in vitro'', the limiting factors are mostly the capacity of a kit versus the maturation time of fluorescent proteins. Since we used a fast-folding GFP variant and mCherry, which was characterized with a maturation time of 15 minutes by Tsien and Coworkers, the problem should be minimized. Alternative tags may be considered, a major advantage of measuring translation ''in vitro'' may be the use of non-cell permeable tags for switch evaluation. |
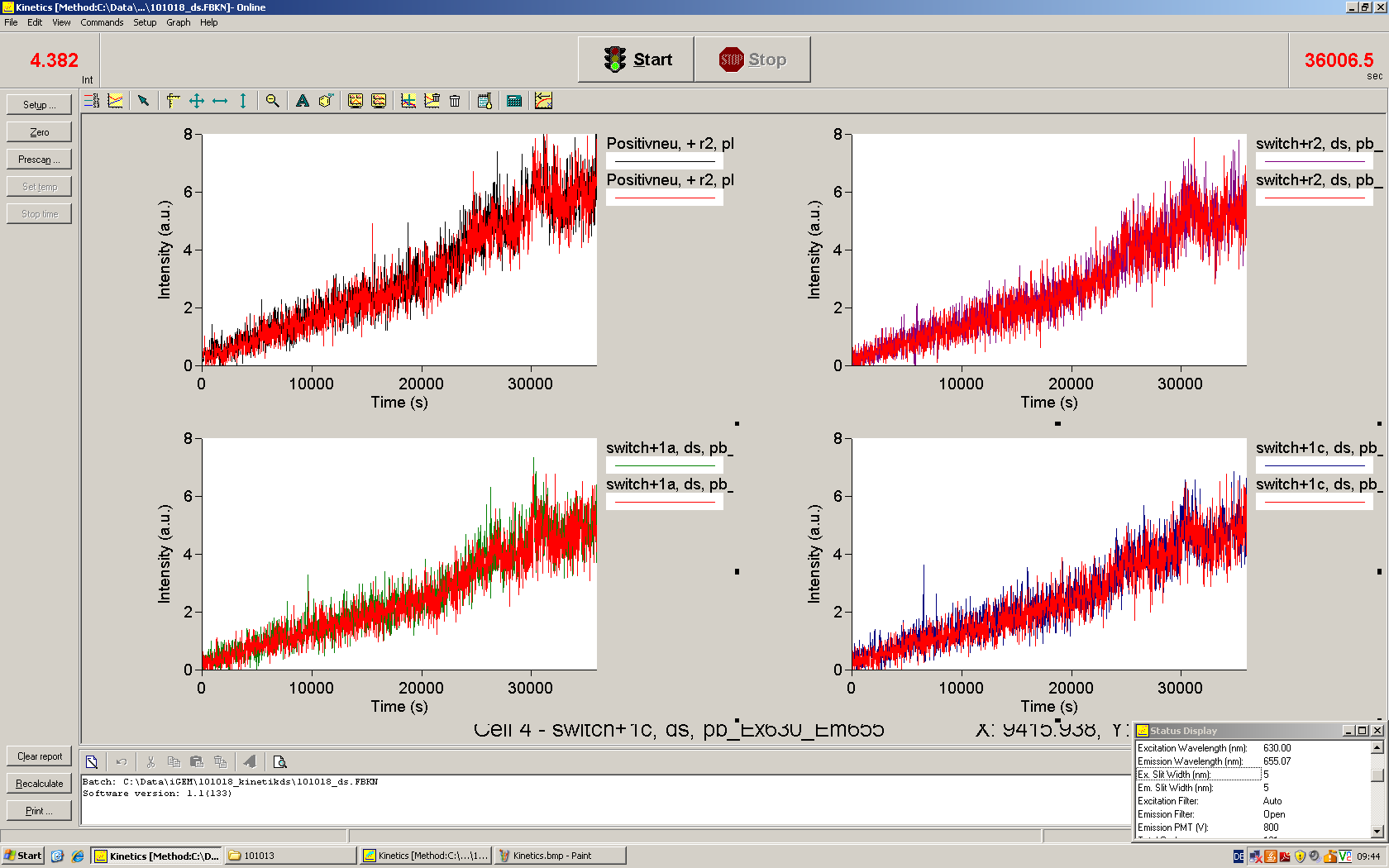
===Measurements=== | ===Measurements=== | ||
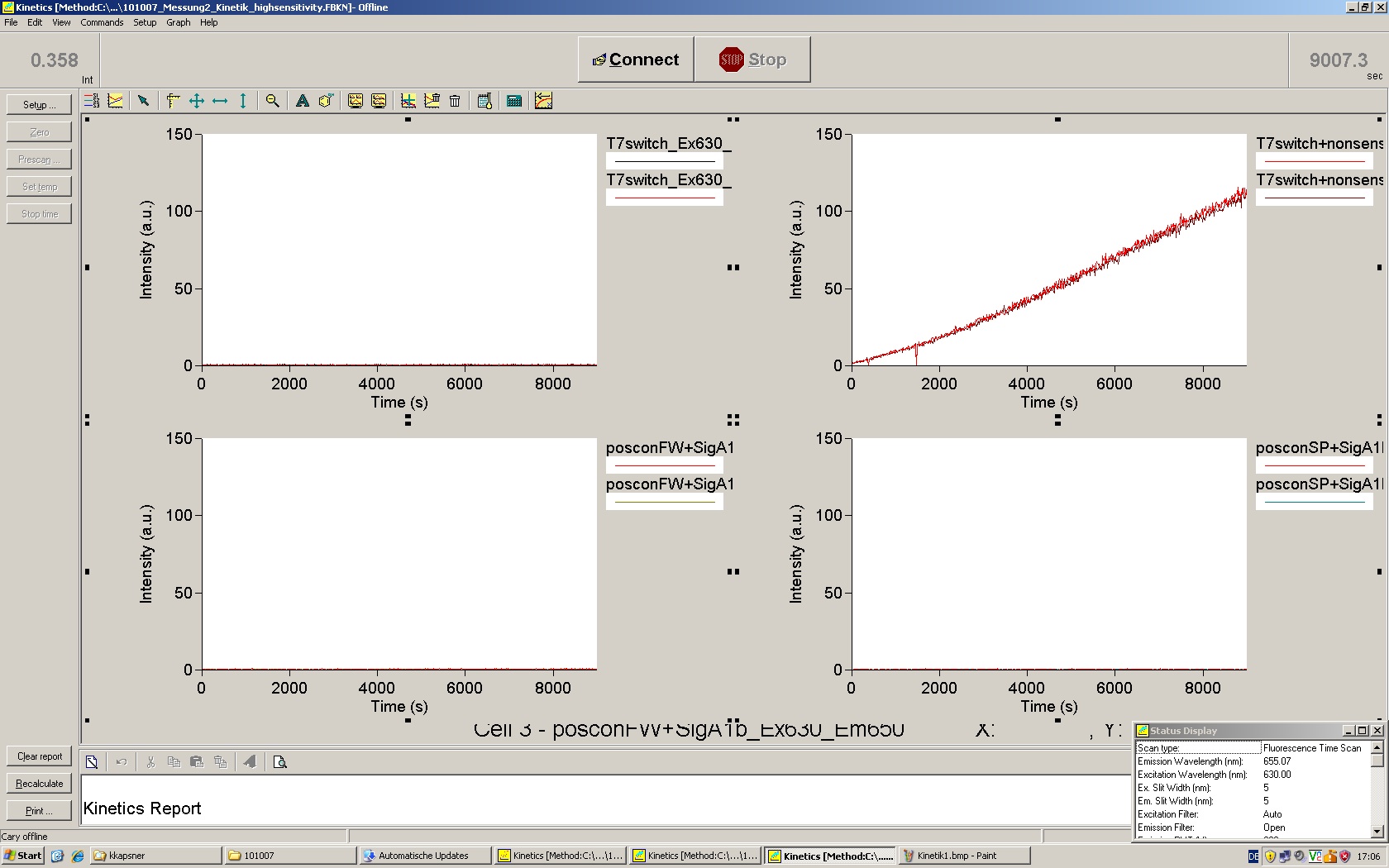
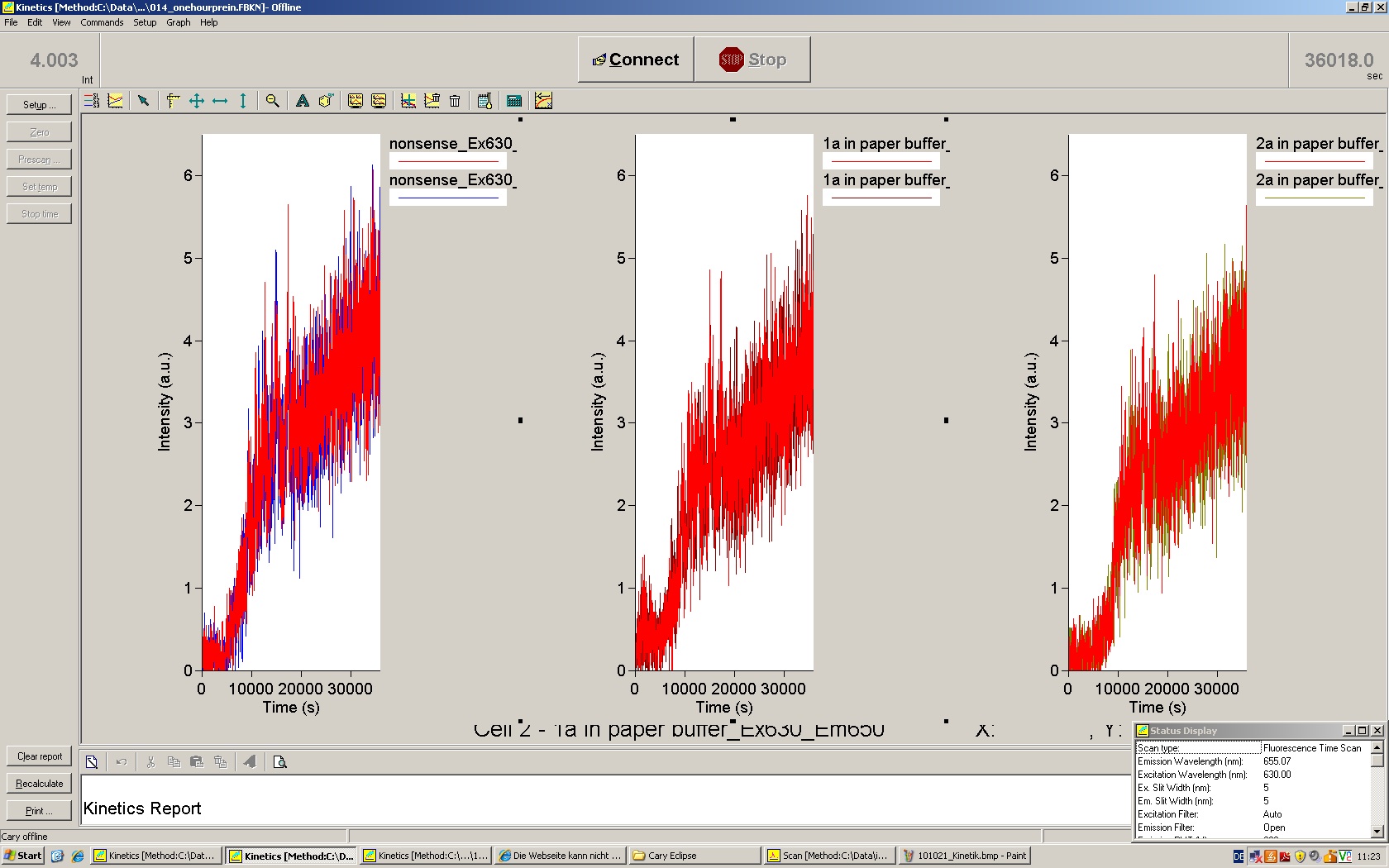
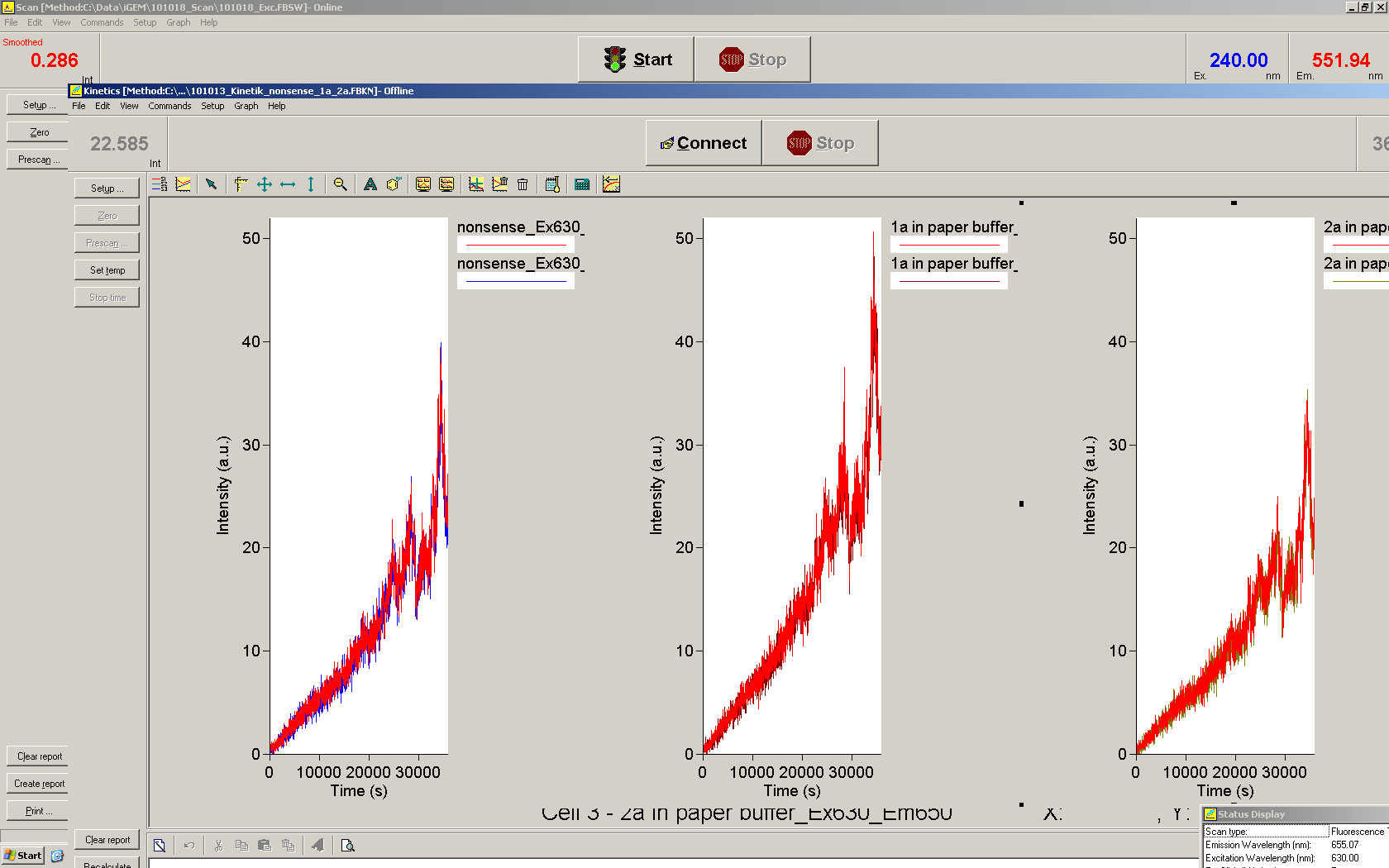
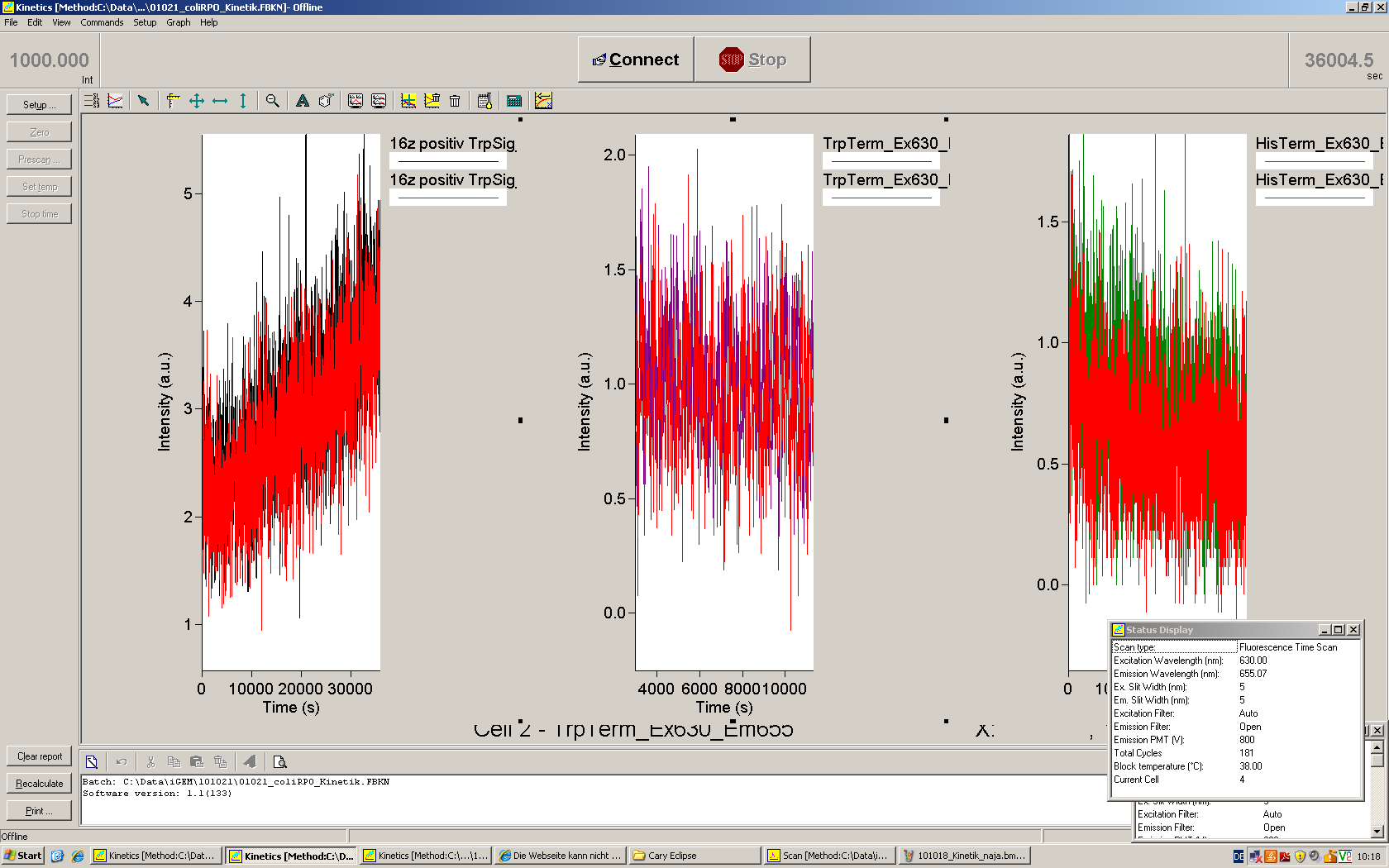
We used the cell-free ''E. coli'' S30 extract system for circular DNA provided by Promega<sup>[[Team:TU_Munich/Lab#ref1|[1]]]</sup>, which is prepared by modifications of the Method Zubay ''et al.''<sup>[[Team:TU_Munich/Lab#ref2|[2]]]</sup>. The characterization of the kit can be obtained from the [http://partsregistry.org/Cell-free_chassis/Commercial_E._coli_S30 Parts Registry]. <br> | We used the cell-free ''E. coli'' S30 extract system for circular DNA provided by Promega<sup>[[Team:TU_Munich/Lab#ref1|[1]]]</sup>, which is prepared by modifications of the Method Zubay ''et al.''<sup>[[Team:TU_Munich/Lab#ref2|[2]]]</sup>. The characterization of the kit can be obtained from the [http://partsregistry.org/Cell-free_chassis/Commercial_E._coli_S30 Parts Registry]. <br> | ||
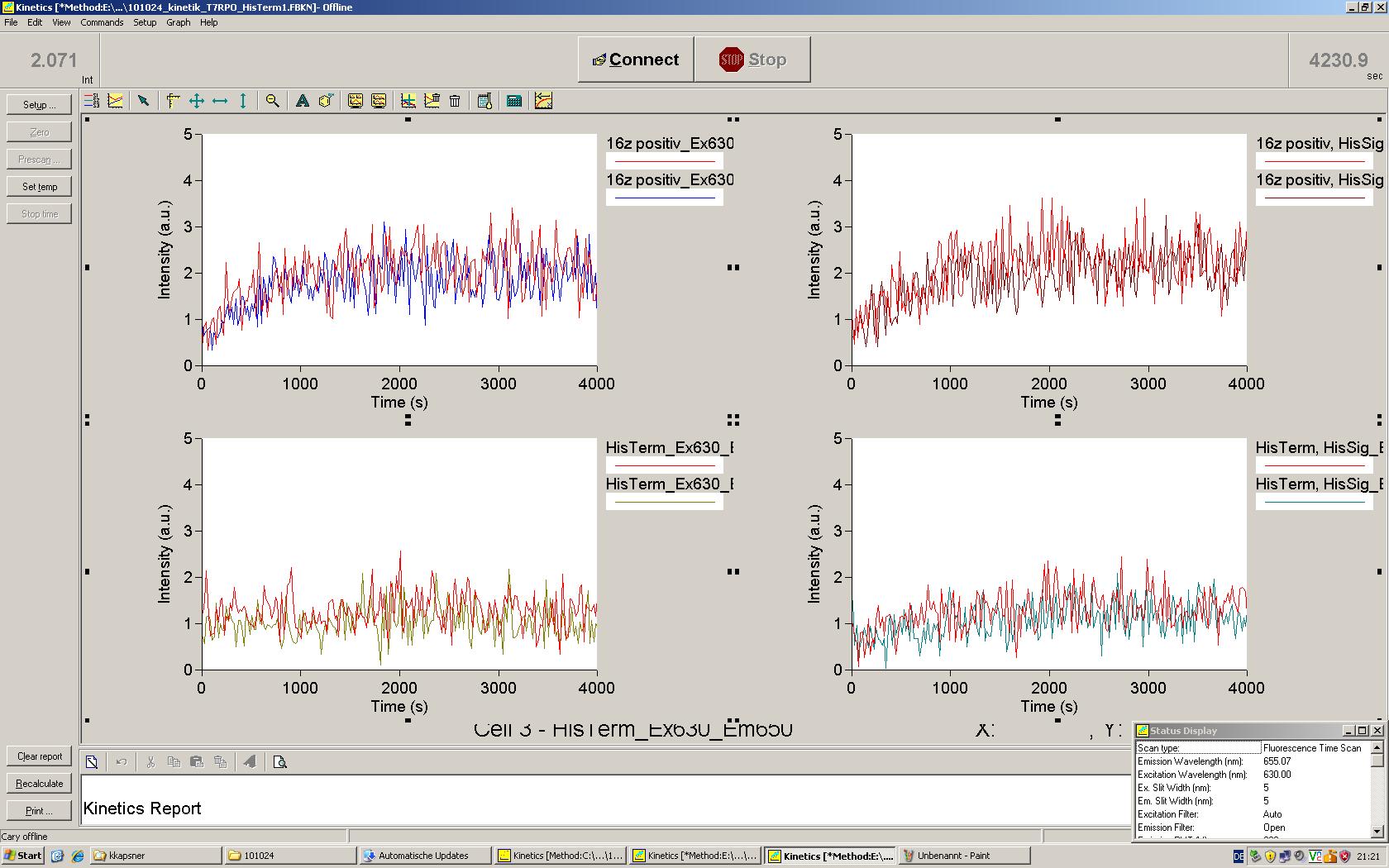
Experiments were performed at 37°C with an amount of approximately 1 µg plasmid in a reaction volume of 50 µL. Fluorescence was followed over time in a jasco fluorolog with wavelength corresponding to those used ''in vivo''. | Experiments were performed at 37°C with an amount of approximately 1 µg plasmid in a reaction volume of 50 µL. Fluorescence was followed over time in a jasco fluorolog with wavelength corresponding to those used ''in vivo''. | ||
| - | |||
| - | |||
| - | |||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
==''In vitro'' Transcription== | ==''In vitro'' Transcription== | ||
| - | To monitor transcription termination and antitermination on a the molecular level, ''in vitro'' transcription of individual switches and their response to signals offer an elegant way for fast and easy prove of principle. Most side effects | + | To monitor transcription termination and antitermination on a the molecular level, ''in vitro'' transcription of individual switches and their response to signals offer an elegant way for fast and easy prove of principle. Most side effects occurring in a complex environment given in a cell or a cell lysate do not arise here. Another major advantage of ''in vitro'' transcription experiments is the possibility to test many signals for one switch to optimize antitermination efficiency and binding specificity without much cloning work. Data gained by ''in vitro'' transcription experiments can be used to improve switches and signals for ''in vivo'' usage. |
{{:Team:TU Munich/Templates/ToggleBoxStart}} | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| - | Since we are working on a totally new principle of | + | Since we are working on a totally new principle of transcriptional control, we used this approach beside the above mentioned advantages for easy variation of different variables like the length of the core unit and the switch to signal ratio. <br> |
| - | To study the switches on a transcriptional level offers the advantage, to reduce interference and possible | + | To study the switches on a transcriptional level offers the advantage, to reduce interference and possible artifacts to a minimum. Since we are not sure how cellular mechanisms like degradation of RNases or interacting factors as well as molecular crowding influence our systems, ''in vitro'' transcription was also used as a minumum system from which more complexity can derive. |
<br> | <br> | ||
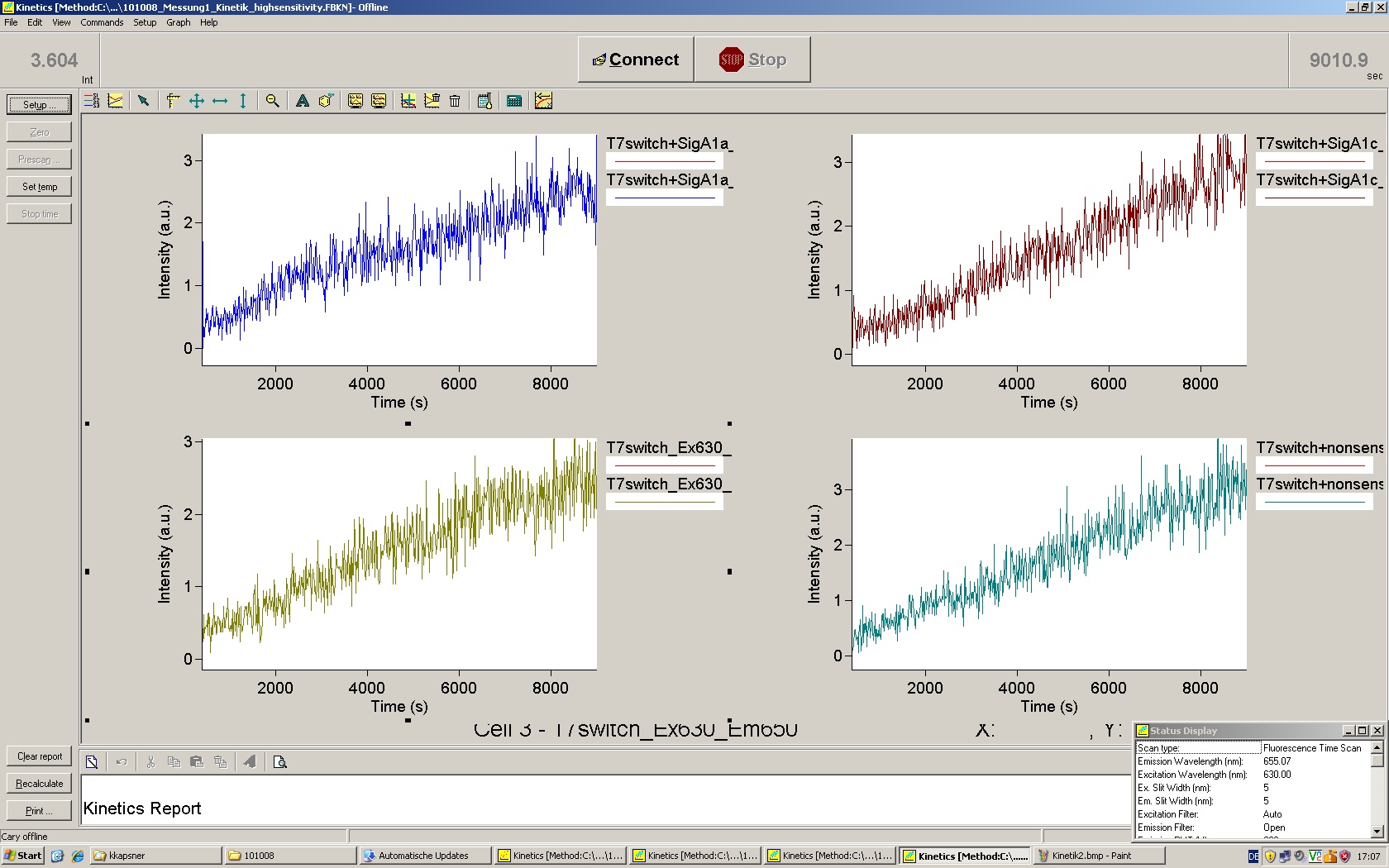
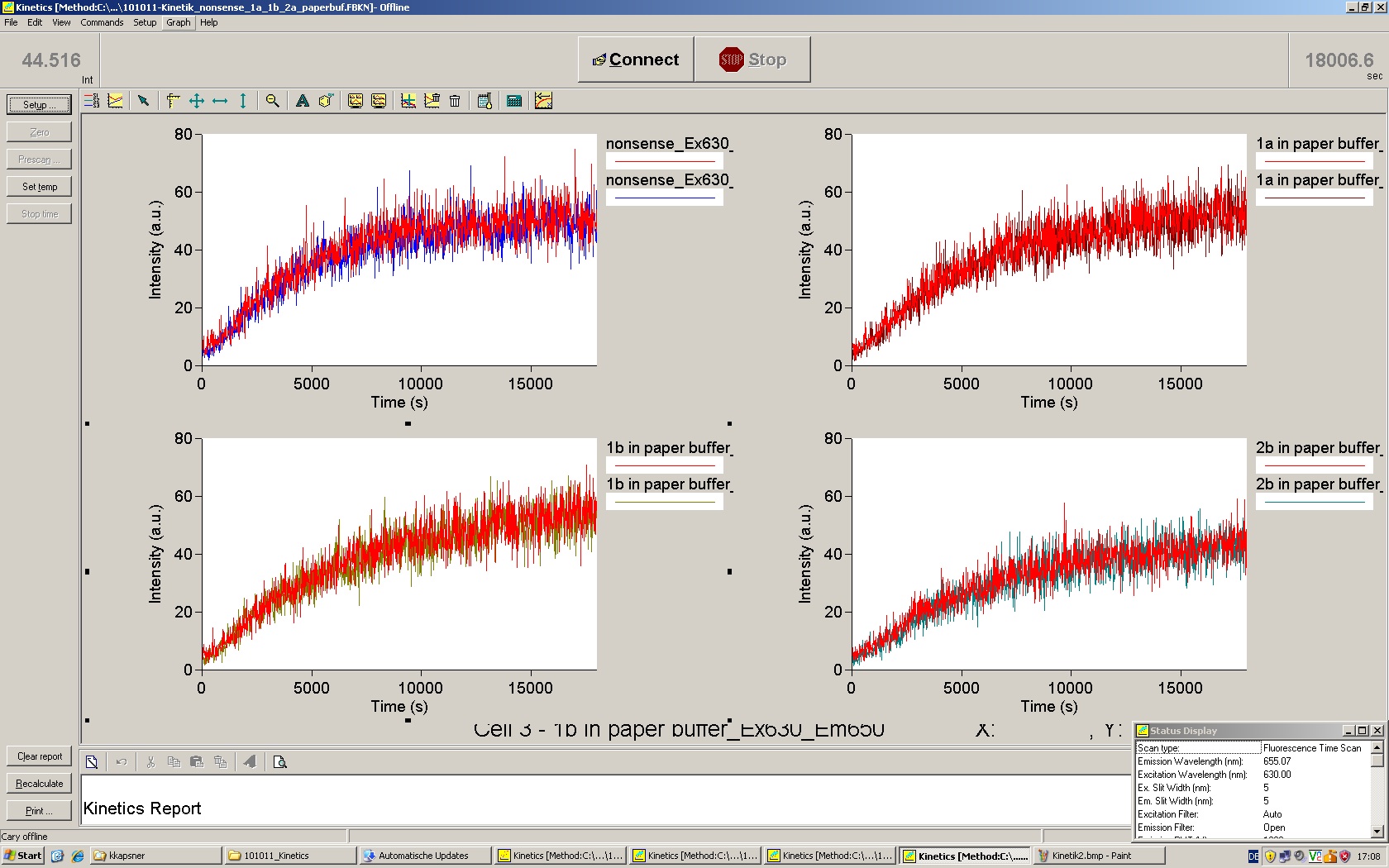
| - | Working with in vitro systems also has the advantage that an input is not needed anymore and the output can also be generated easily. We used '''two readouts''' with '''two different transcription systems''' to check and investigate our devices: First, we used an [https://2010.igem.org/Team:TU_Munich/Parts malachitegreen-binding aptamer] for an | + | Working with in vitro systems also has the advantage that an input is not needed anymore and the output can also be generated easily. We used '''two readouts''' with '''two different transcription systems''' to check and investigate our devices: First, we used an [https://2010.igem.org/Team:TU_Munich/Parts malachitegreen-binding aptamer] for an fluorescence output and second, we simply put our reaction educts on an denaturing acrylamide-gel to check for RNA varying in length. As for two different transcription systems we used on the one hand ''E. coli''-RNA Polymerase (RPO) based transcription since the aim is to apply the so gained results ''in vivo'' and on the other hand T7 based transcription which is well established through literature and delivers good RNA yields. |
<br> | <br> | ||
=== T7 RNA polymerase === | === T7 RNA polymerase === | ||
| Line 87: | Line 89: | ||
<div align="justify"> | <div align="justify"> | ||
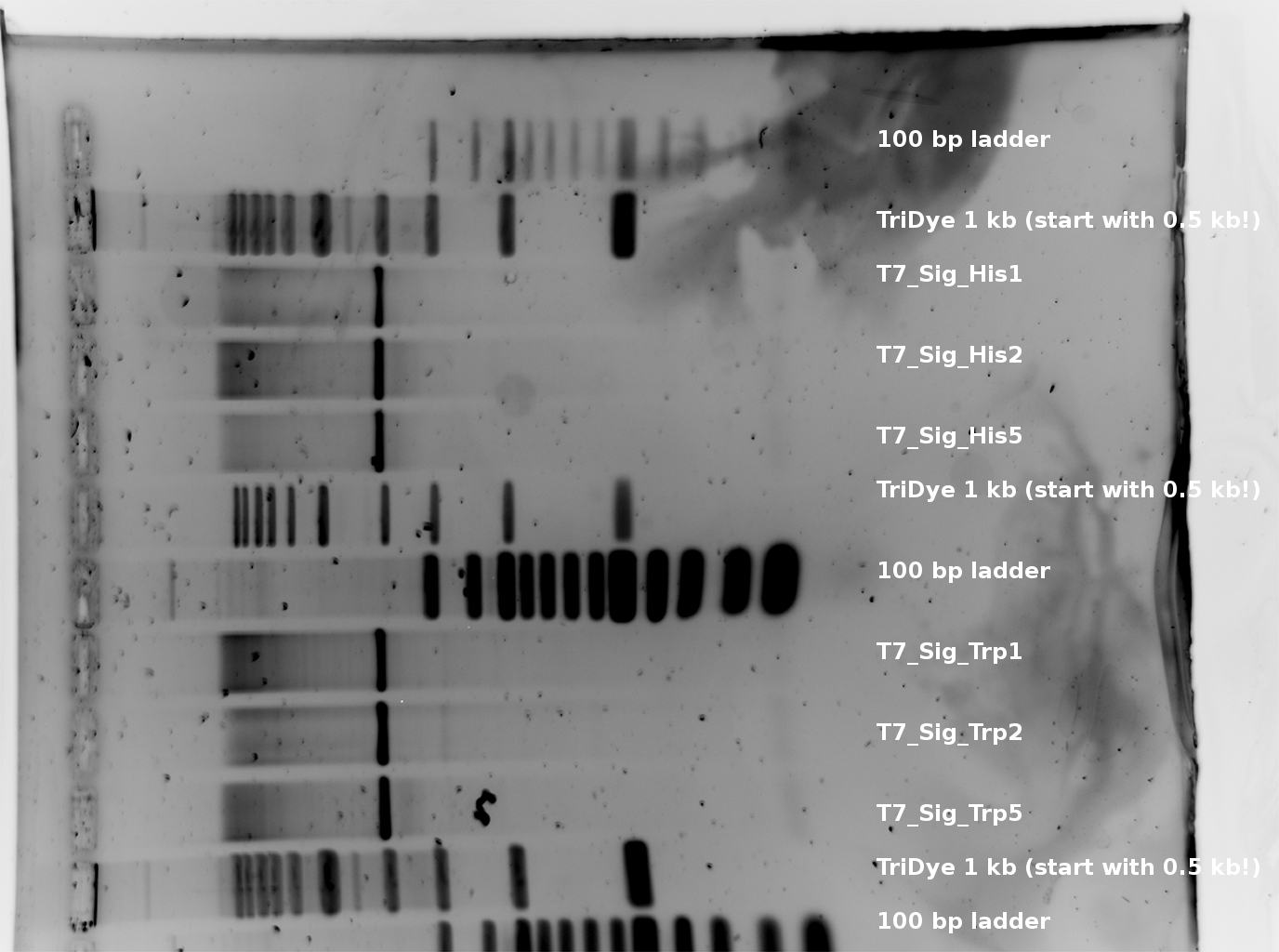
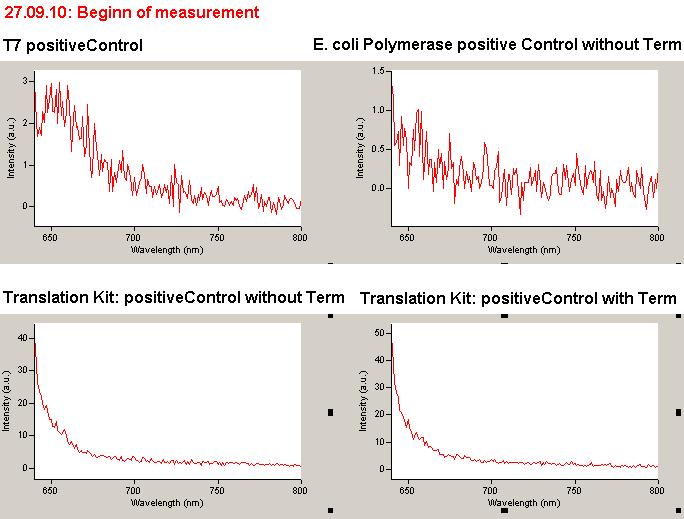
| - | The T7 RNA polymerase is known for satisfying RNA yields together with easy handling. In our approach we had PCR amplified, double stranded switches with an malachitegreen binding aptamer following after | + | The T7 RNA polymerase is known for satisfying RNA yields together with easy handling. In our approach we had PCR amplified, double stranded switches with an malachitegreen binding aptamer following after a T7 terminator which was constructed to function as a switch. Different signals were tested varying in length of the specificity site and the triggering unit. |
| - | + | ||
<br> | <br> | ||
| + | For in vitro expression the T7 RNA Polymerase requires a double stranded promotor region at the beginning of the DNA template but is otherwise capable of handling single stranded DNA, so a sense strain corresponding to the T7 promoter region was added. Transcription is more effective with double stranded DNA as template. Apart from that, no more requirements are needed in theory which makes the evaluation of many signals especially easy. Since we ordered the signal sequences we tested we chose the cheaper way in the beginning by using single stranded signals with corresponding sense T7 pieces and switched to double stranded constructs after narrowing down the most promising switch/signal pairs. Later on we also used double stranded signals and switches since transcription rates are higher with those. | ||
| + | <br> | ||
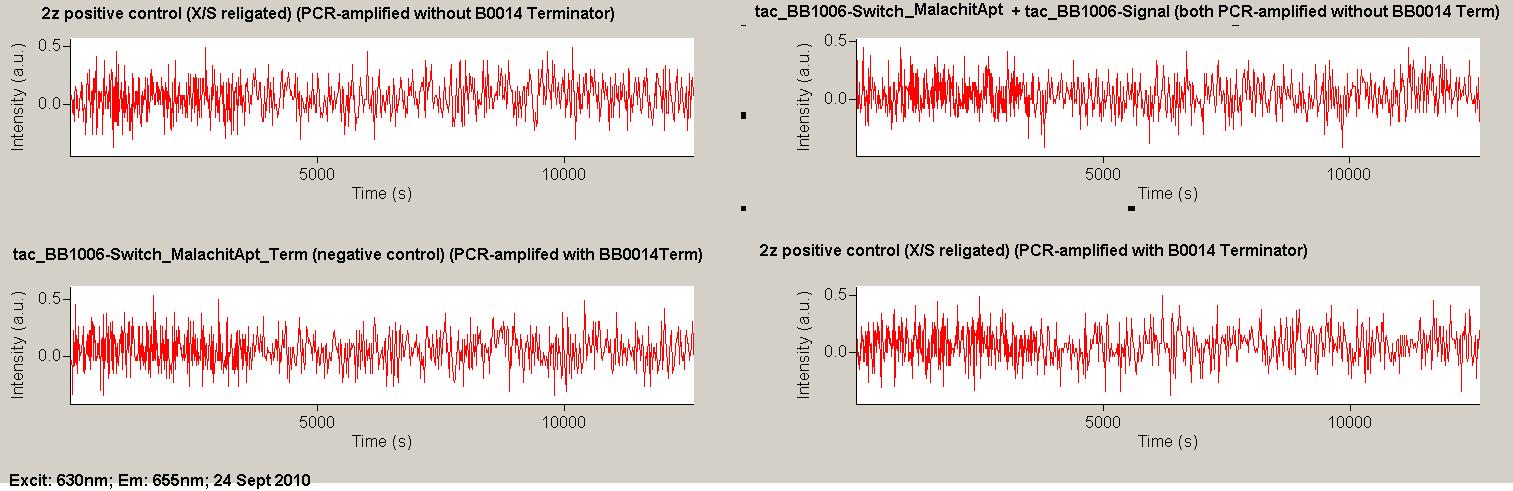
| + | As a positive control, the malachite green binding aptamer right behind the T7 promoter was used. Transcription proceeds without termination and the maximal fluorescence intensity should be gained. | ||
| + | <br> Transcription termination can also be estimated by measuring just the switch without interfering signals. Since upon transcription of a signal sequence, less RNA Polymerase is available, the transcription rate of the switch and therefore the fluorescence output is reduced by merely adding the signal. Therefore randomly chosen short sequences in the range of the tested signals were added to the negative control. | ||
| - | |||
</div> | </div> | ||
| - | |||
| - | |||
=== ''E. coli'' RNA polymerase === | === ''E. coli'' RNA polymerase === | ||
| - | In comparison to the T7 RNA Polymerase the ''E. coli'' RNA Polymerase requires slightly more sophisticated proceedings when it comes to the design of switches and handling of the enzyme. The biggest in our case was to store it properly since the only -80°C fridge was in another building. <br> | + | In comparison to the T7 RNA Polymerase the ''E. coli'' RNA Polymerase requires slightly more sophisticated proceedings when it comes to the design of switches and handling of the enzyme. The biggest in our case was to store it properly since the only -80°C fridge was in another building, so make sure you have a big supply of dry ice ready if you encounter the same problem. <br> |
| - | E. coli RPO was ordered saturated with σ70-factor | + | E. coli RPO was ordered saturated with σ70-factor. |
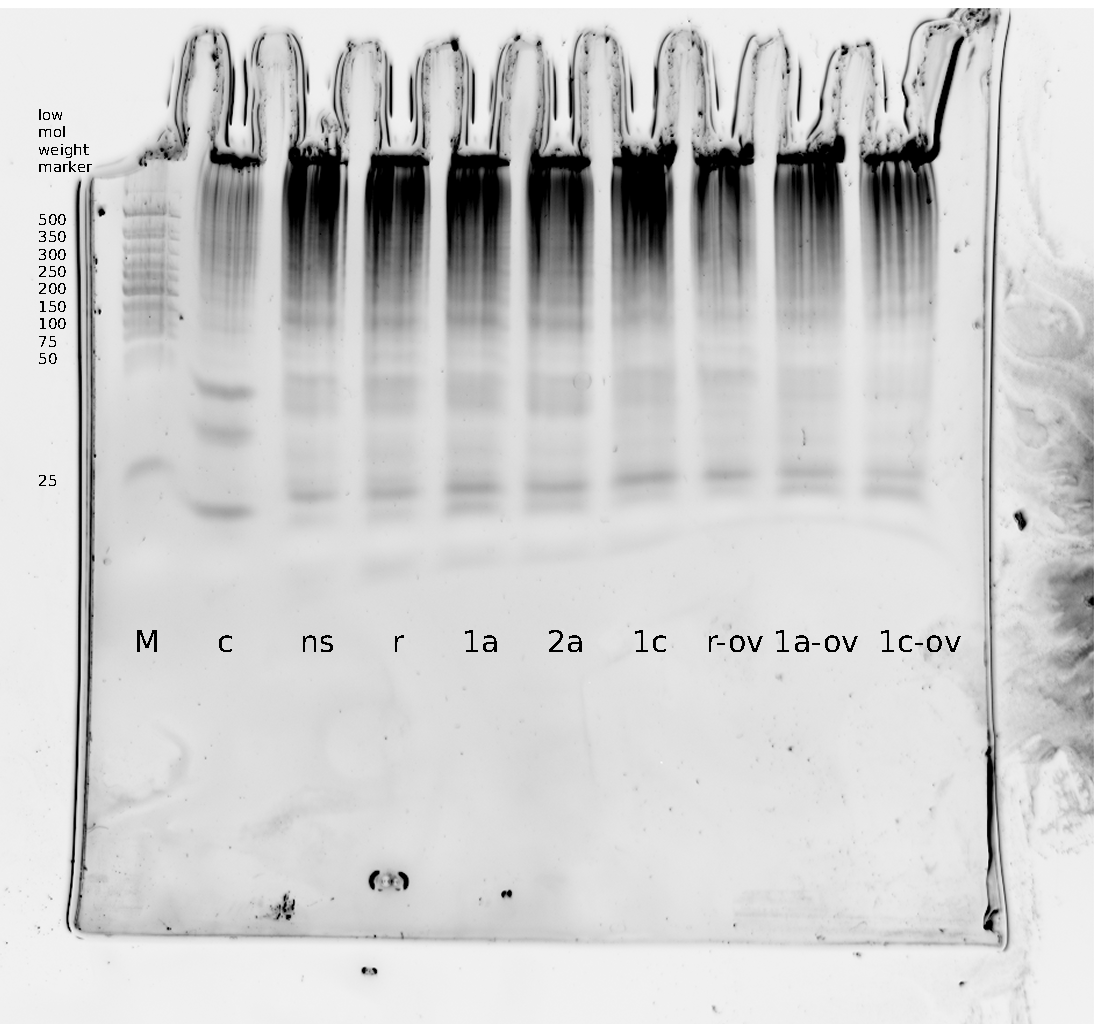
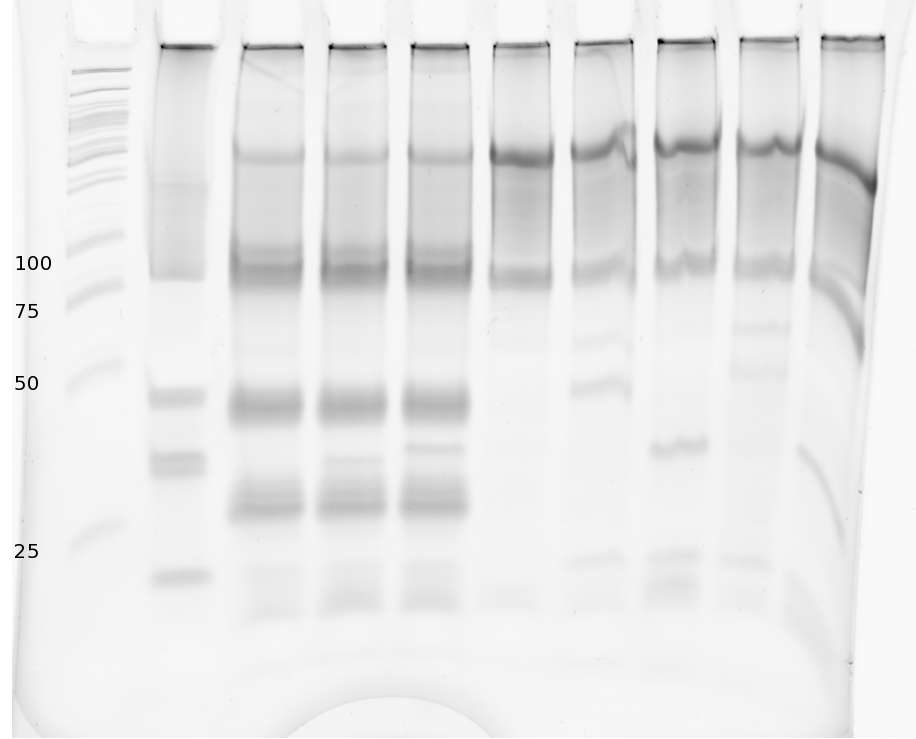
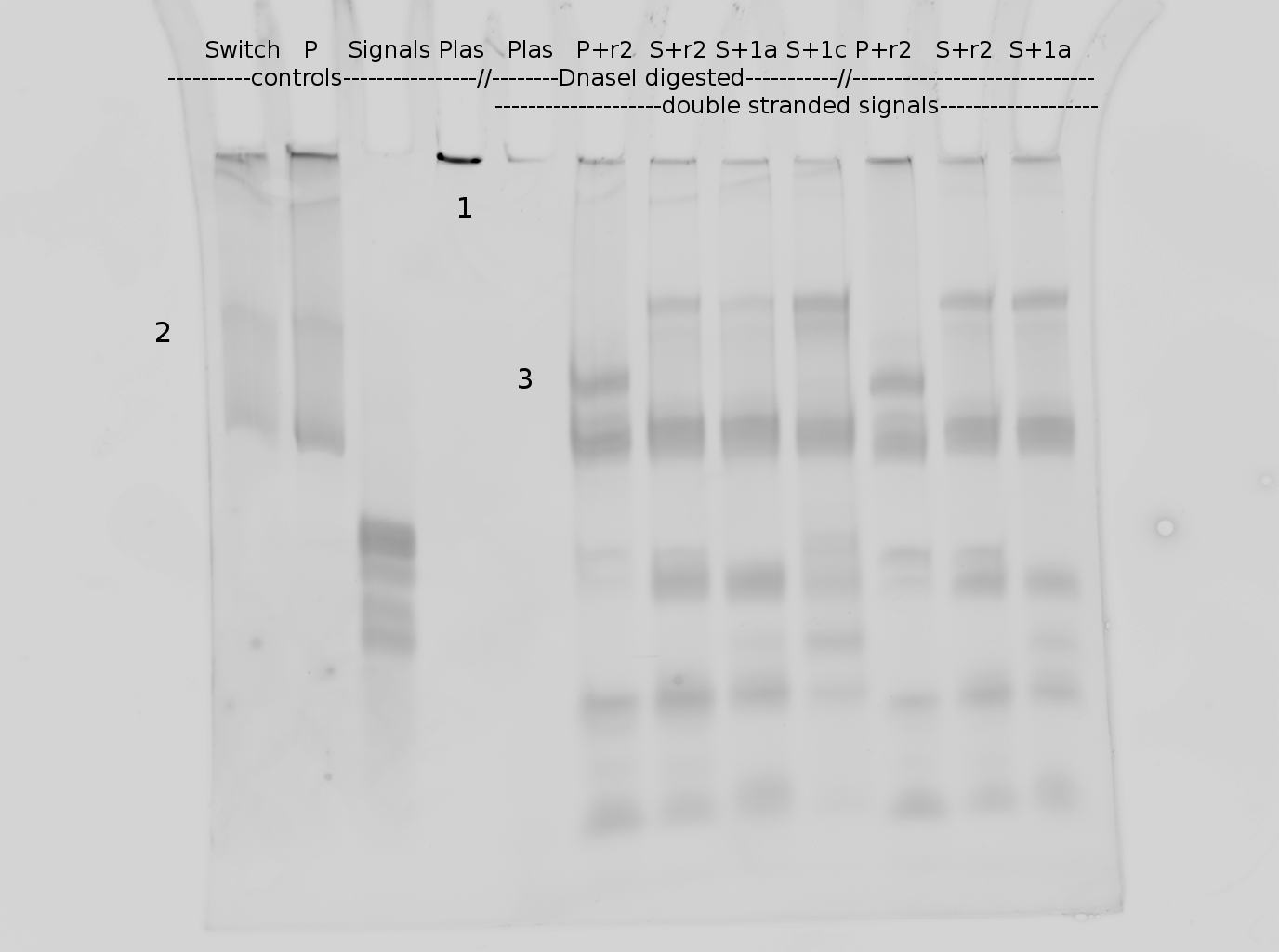
=== Denaturing Polyacrylamide gel electrophoresis === | === Denaturing Polyacrylamide gel electrophoresis === | ||
<div align="justify"> | <div align="justify"> | ||
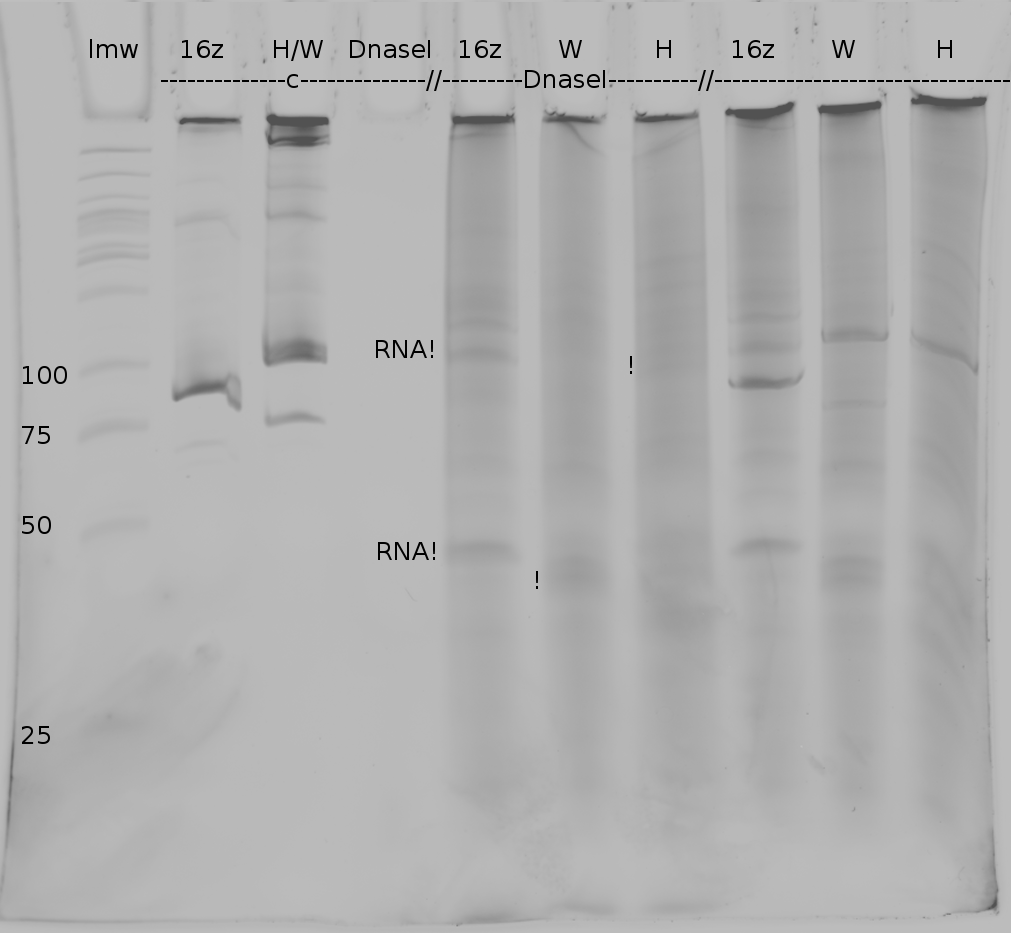
| - | + | Polyacrylamide gel electrophoresis (PAGE) was used for evaluation of termination and switching efficiency. Gels containing 15 % acrylamide and 6 M urea were used for separation of terminated and readthrough RNAs. The same constructs as designed for the malachite green binding aptamer were used. | |
| - | + | ||
<br> | <br> | ||
| - | Polyacrylamide gels | + | Polyacrylamide gels separate RNA and DNA according to their size in an electric field. Since the negative charge equals the size of nucleotides in the RNA/DNA, the number of base pairs can be compared between two samples often with one base pair resolution. Since RNA forms three-dimensional structures, the samples are preheated and run in 6 or 7 M urea. The polyacrylamide gel is stained in SybrGold afterwards which binds to both single and double stranded DNA and RNA. A Dnase digestion was applied before running the samples to avoid confusion caused by DNA templates. |
<br> | <br> | ||
| - | Denaturing PAGE is a simple yet elegant way to check for transcription efficiency and termination rates. Since it is a very direct way and it provides a simple yet clear readout, we used it as another method beside the more sophisticated malachitegreen binding assay to evaluate and characterize our switch. | + | Denaturing PAGE is a simple yet elegant way to check for transcription efficiency and termination rates. Since it is a very direct way and it provides a simple yet clear readout, we used it as another method beside the more sophisticated malachitegreen binding assay to evaluate and characterize our switch. Equipment for denaturing PAGE can be found in nearly every biochemical lab, so this method also applies for an easy controlexperiment. |
</div> | </div> | ||
| Line 115: | Line 116: | ||
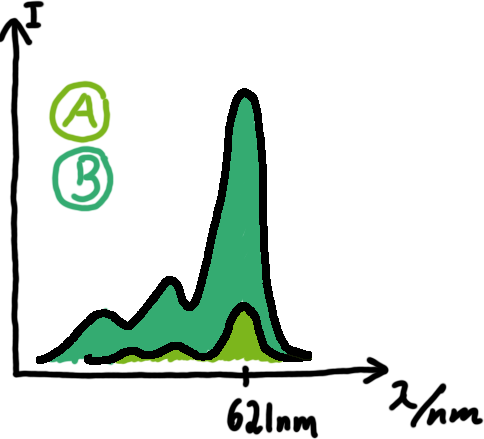
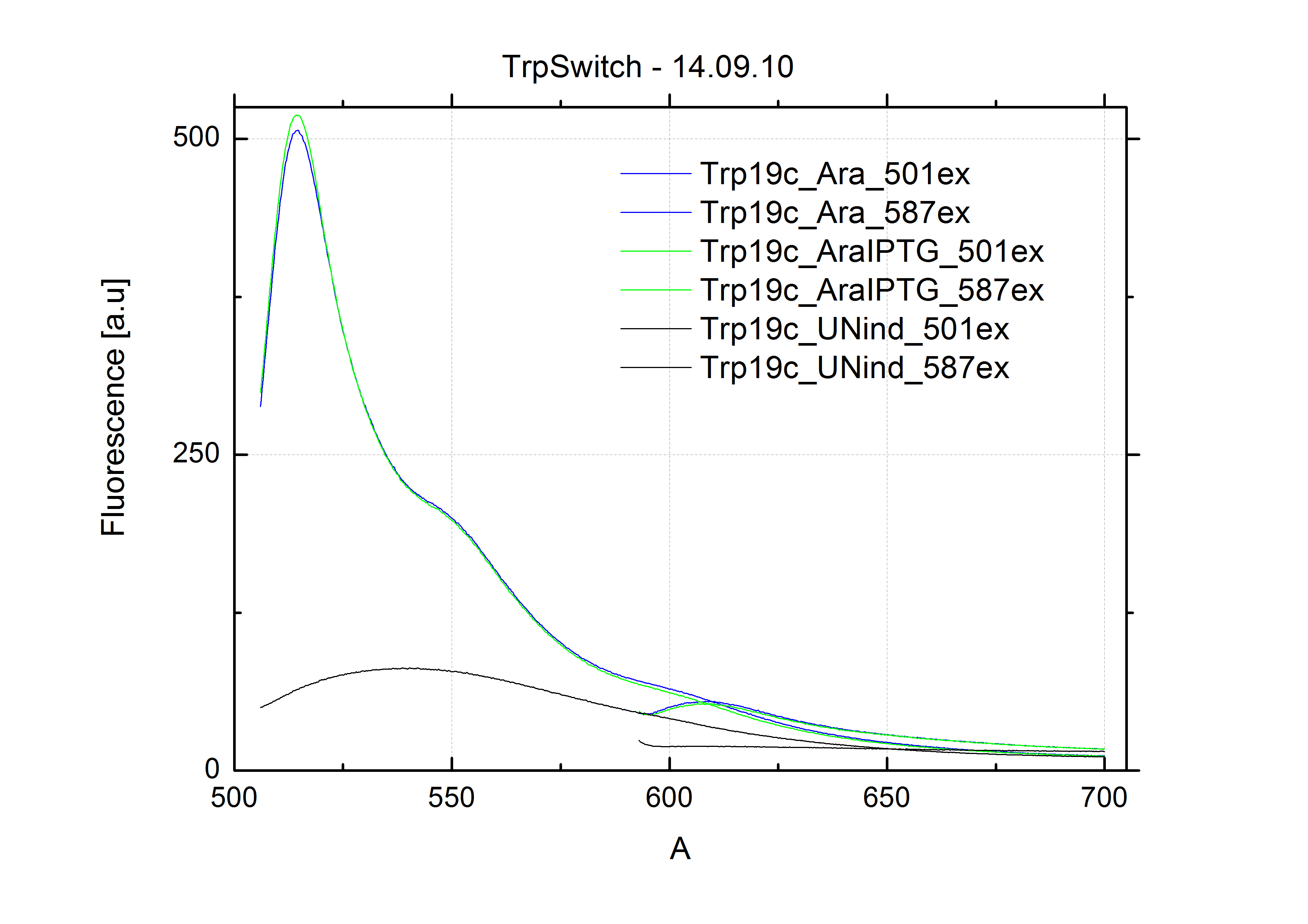
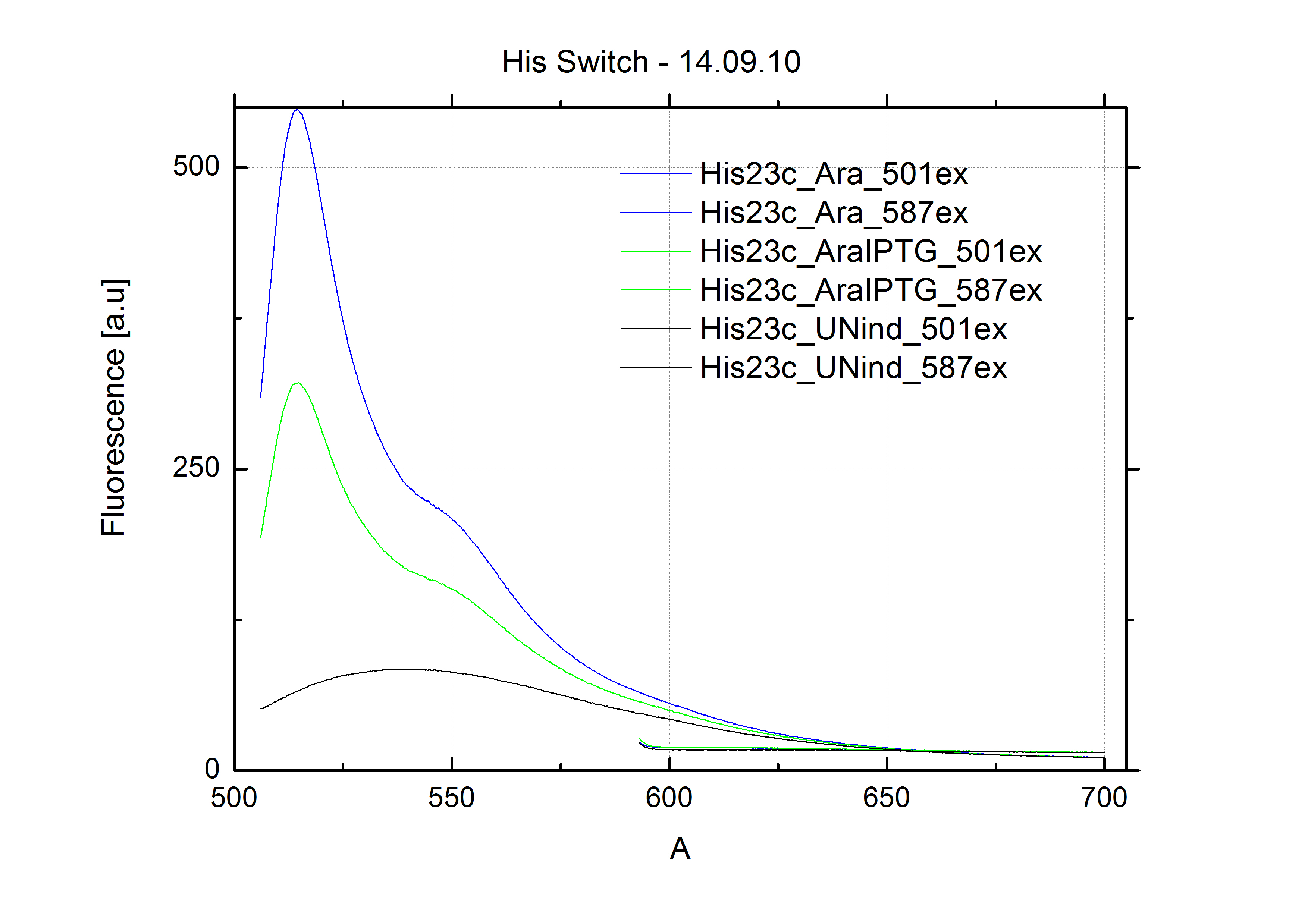
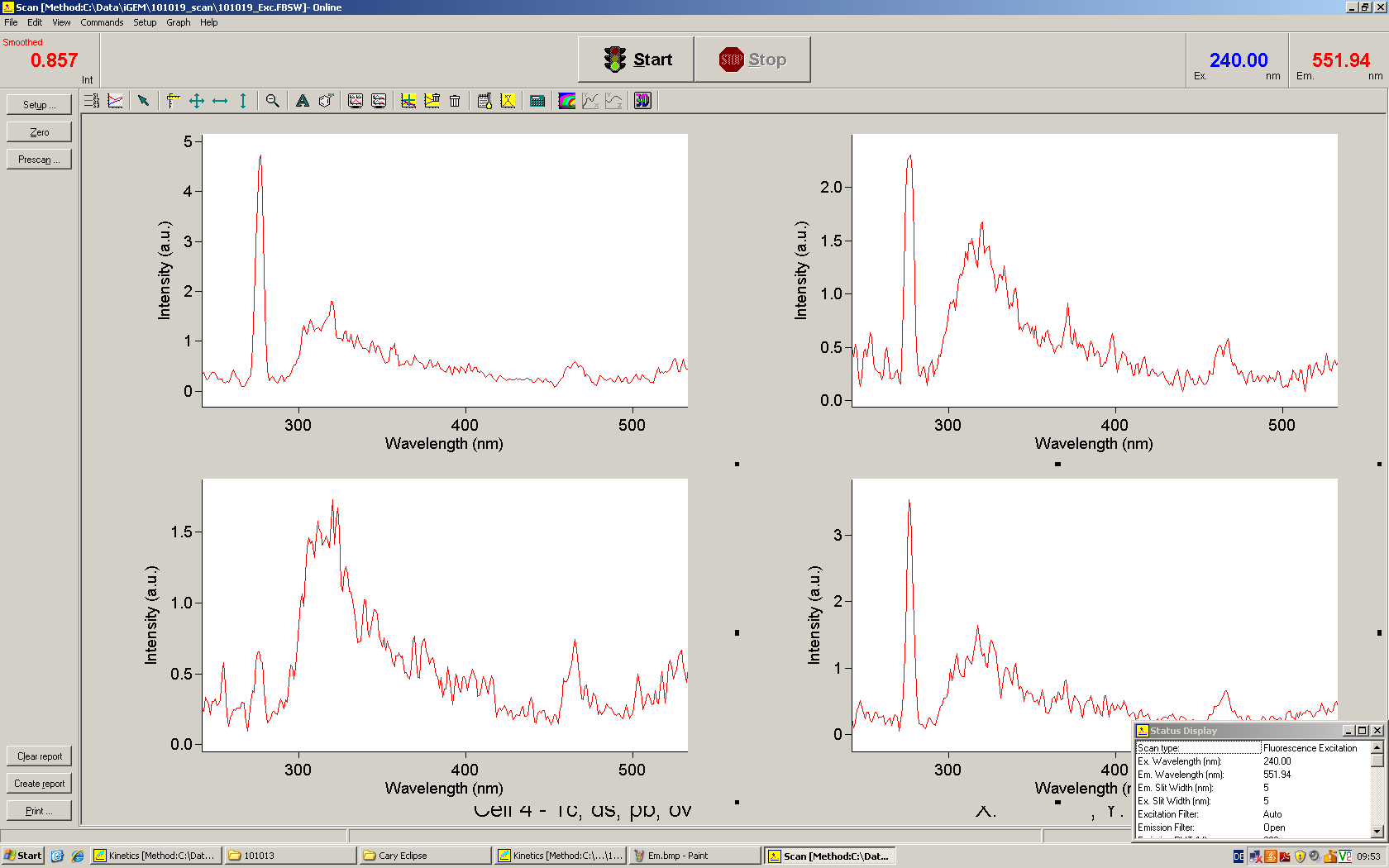
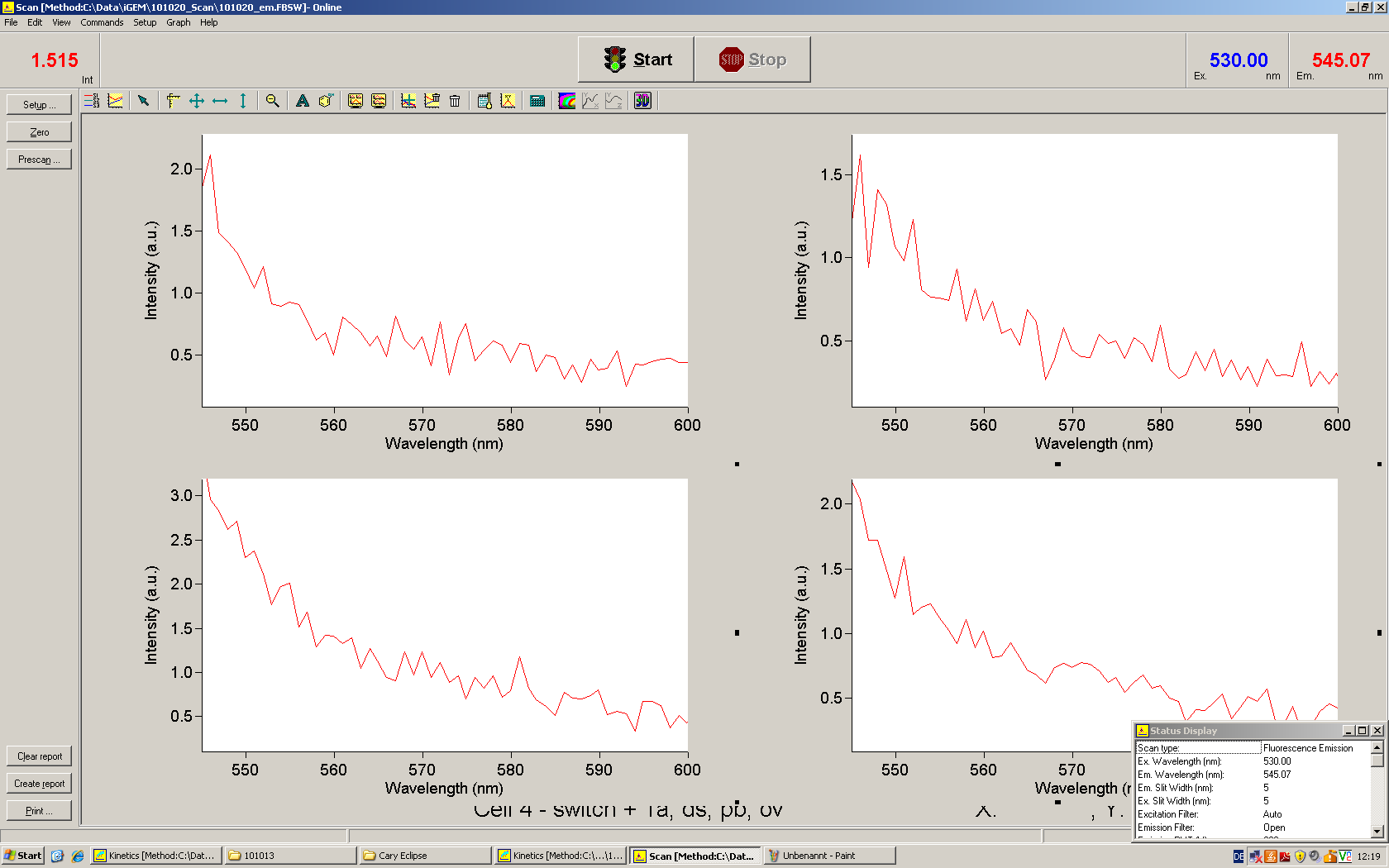
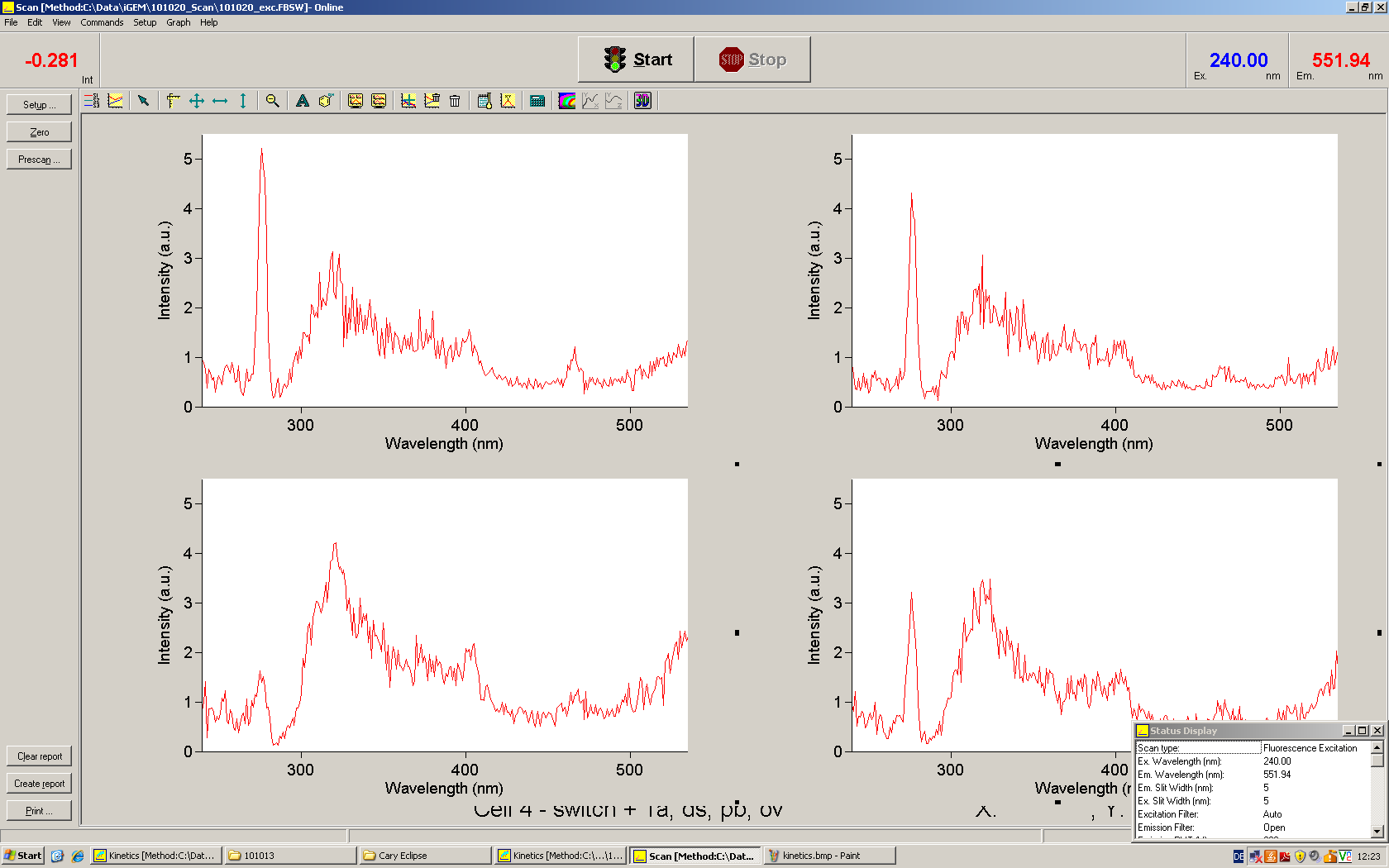
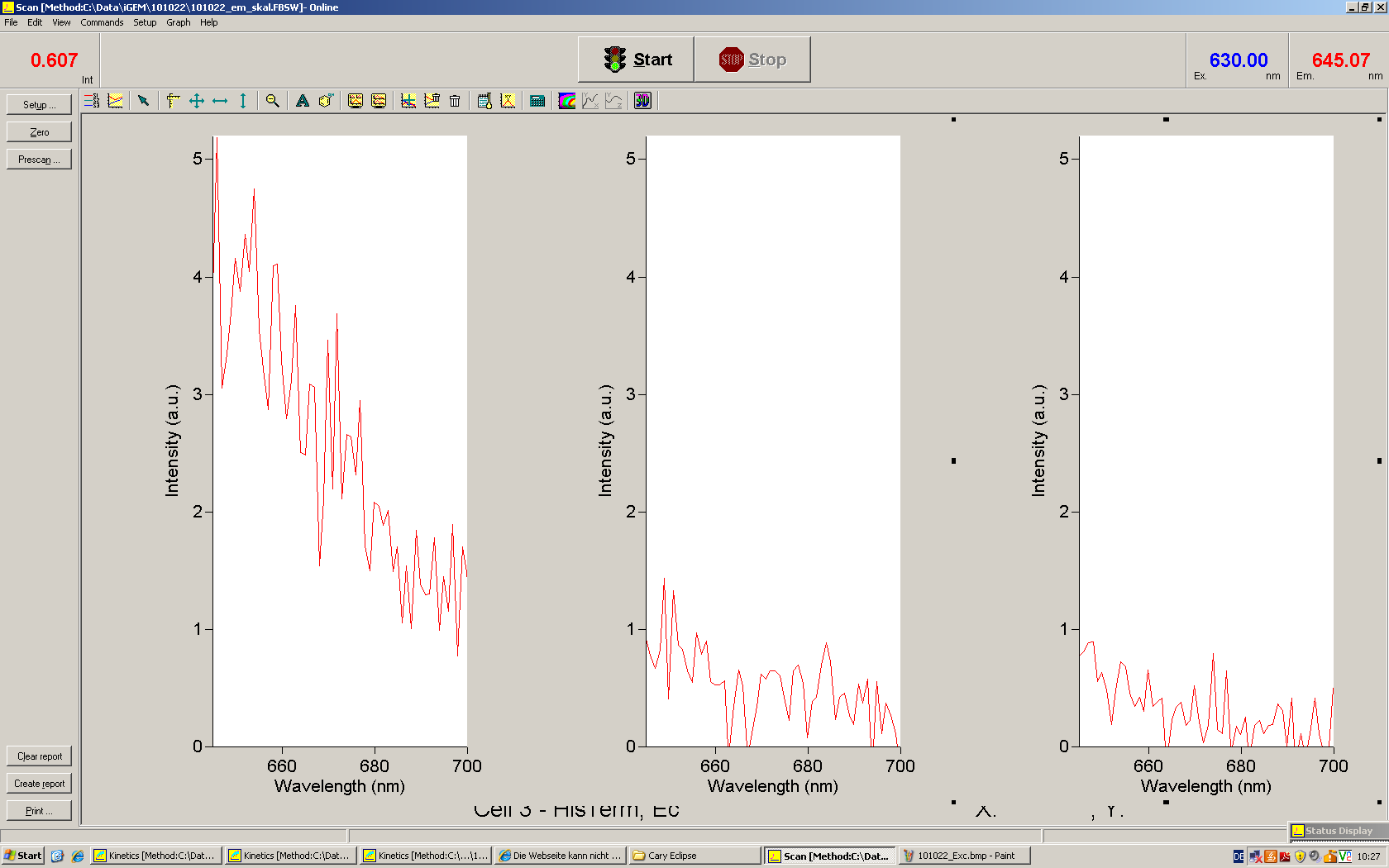
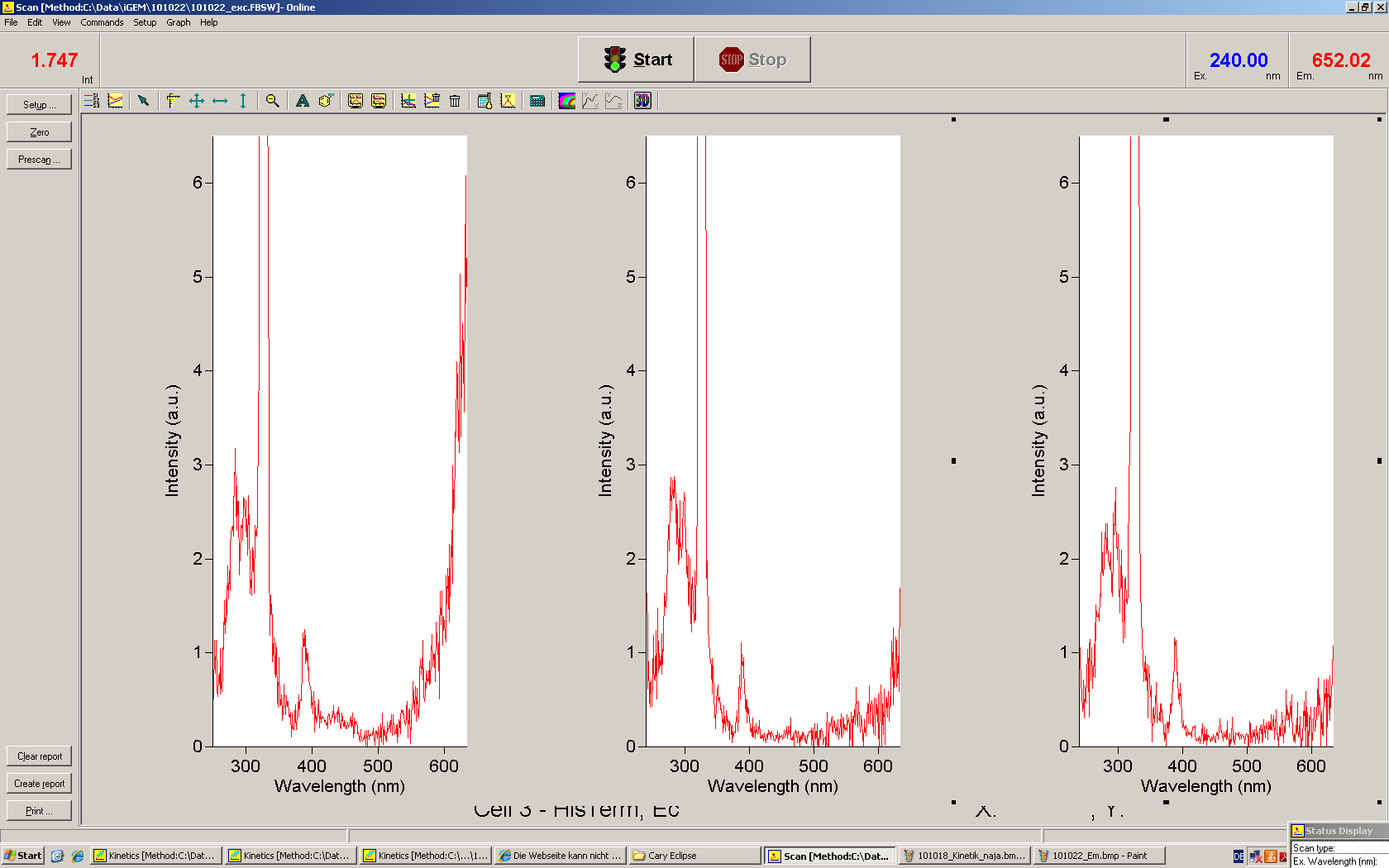
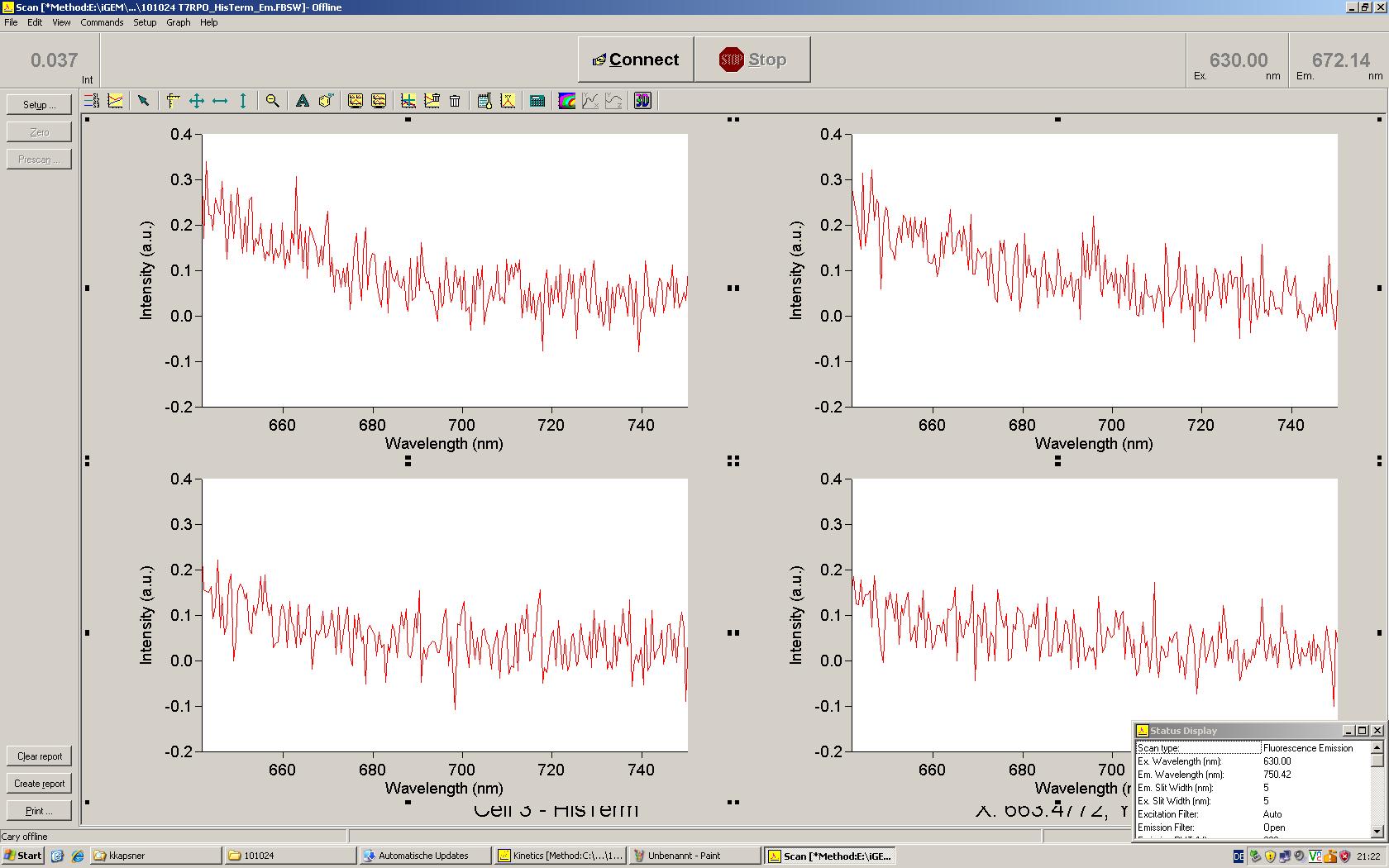
[[Image:TUM2010_Malachit_emission.png|200px|thumb|left|Emission spectra of malachite green; A: without signal-RNA, B: with signal-RNA]] | [[Image:TUM2010_Malachit_emission.png|200px|thumb|left|Emission spectra of malachite green; A: without signal-RNA, B: with signal-RNA]] | ||
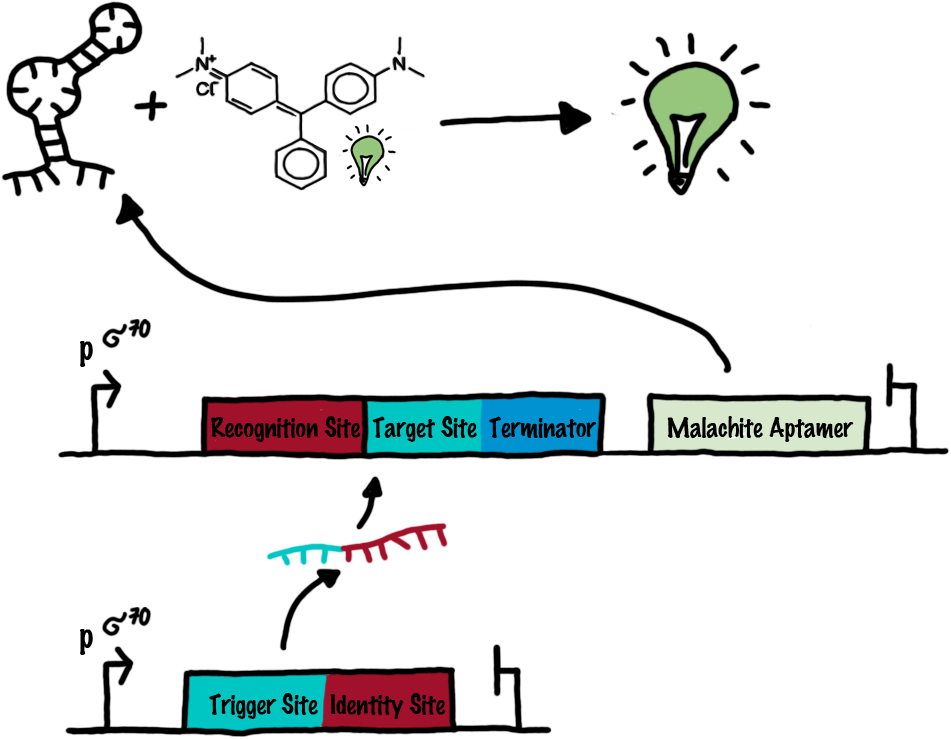
| - | + | In this year's DNA submission we contribute the [[Team:TU_Munich/Parts#Malachite_Green_Binding_Aptamer_-_BBa_K494000 | malachite green binding aptamer]] which can be used as a transcription reporter in ''in vitro'' transcription experiments. <br> | |
| - | <br> | + | Malachite green is a dye with a negligible fluorescence in solution but undergoes a dramatic increase about 3000 times if bound by the RNA aptamer making it an exceptional good marker. Since the binding is very specific, transcription in dependence of a signal can be monitored by measuring the fluorescence of malachite-green over time if the aptamer is located behind the switch. Transcription of the aptamer will only take place after anti-termination by a signal. An increase should be visible over time. Other triphenyldyes are also recognized with weaker effects on the fluorescence but may also serve as reporters if the emission or excitation of malachite green does not fit the experimental setup. |
| + | <br><br><br> | ||
| + | [[Image:TUM2010_Malachitgruen-2.png|600px|center|thumb|Description of the malachit green assay. Antitermination allows the polymerase to produce the malachite green aptamer ]] | ||
| + | Malachite green binding can be used to follow RNA transcription over time, a rise in the fluorescence is then detectable. Fluorescence marker of specific RNA structures are still rare, so the malachite green binding aptamer provides one of the only possibilities to continuously monitor transcription reactions. In comparison to PAGE, kinetics can be taken, while with PAGE only end point estimations can be made. This makes the malachite green binding aptamer a valuable tool to study ''in vitro'' transcription in general and the principles underlying our switch in principle. | ||
| + | |||
| + | |||
| + | <!--For the T7-based measurements we ordered single stranded signals for a first attempt and added matching single strands complementary to the T7 promoter region. The switch was amplified using PCR and consisted of the following elements: Primer-binding site - T7 promotor - switch - malachitegreen binding aptamer. Upon binding of a correct signal to the switch, the stem loop dissolves and transcription is possible. | ||
| - | |||
<br><br> | <br><br> | ||
| - | OLD: A second possibility to measure parameters of our switches we came up with, was the idea to investigate our system on the transcriptional level only. Therefore, we decided to use malachite green as reporter. Malachite green in a fluorescent dye, whose emission increasing | + | OLD: A second possibility to measure parameters of our switches we came up with, was the idea to investigate our system on the transcriptional level only. Therefore, we decided to use malachite green as reporter. Malachite green in a fluorescent dye, whose emission increasing dramatically (about 3000 times) upon binding of a specific RNA-aptamer. The RNA-aptamer |
<br><br> | <br><br> | ||
---concept to be described, as well as literature--- | ---concept to be described, as well as literature--- | ||
| Line 131: | Line 137: | ||
We made constructs comprising of a sigma(70)-binding promoter followed by a short nonsense sequence, the switches and the aptamer sequence. | We made constructs comprising of a sigma(70)-binding promoter followed by a short nonsense sequence, the switches and the aptamer sequence. | ||
<br> | <br> | ||
| - | Also we made constructs, where the transcription of the signal-RNA is under the control of a sigma(70) promoter. These two linear DNA-constructs, together with the e.coli RNA-polymerase and the right buffer conditions should represent an easy-to-handle measurement kit on the transcriptional level. | + | Also we made constructs, where the transcription of the signal-RNA is under the control of a sigma(70) promoter. These two linear DNA-constructs, together with the e.coli RNA-polymerase and the right buffer conditions should represent an easy-to-handle measurement kit on the transcriptional level. --> |
| - | </div> | + | </div> |
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| - | = | + | =Lab Book= |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Explanations== | ==Explanations== | ||
In the following we present an overview regarding our work in the lab. For easier understanding we summarized the work of each week using colored boxes. To get a better overview we used the following color code for the boxes: | In the following we present an overview regarding our work in the lab. For easier understanding we summarized the work of each week using colored boxes. To get a better overview we used the following color code for the boxes: | ||
{| | {| | ||
|- | |- | ||
| - | | {{:Team:TU_Munich/Templates/RedBox | text= }} The red box represents general cloning steps that were required for our measurements. See the [[Team:TU_Munich/Lab# | + | | {{:Team:TU_Munich/Templates/RedBox | text= }} The red box represents general cloning steps that were required for our measurements. See the [[Team:TU_Munich/Lab#Molecular_Biology | protocol section]] for further details. |
|- | |- | ||
| {{:Team:TU_Munich/Templates/BlueBox | text= }} The blue box indicates <i>in vivo</i> measurements which are described [[Team:TU_Munich/Lab#In vivo Measurements | here]]. | | {{:Team:TU_Munich/Templates/BlueBox | text= }} The blue box indicates <i>in vivo</i> measurements which are described [[Team:TU_Munich/Lab#In vivo Measurements | here]]. | ||
| Line 614: | Line 189: | ||
---- | ---- | ||
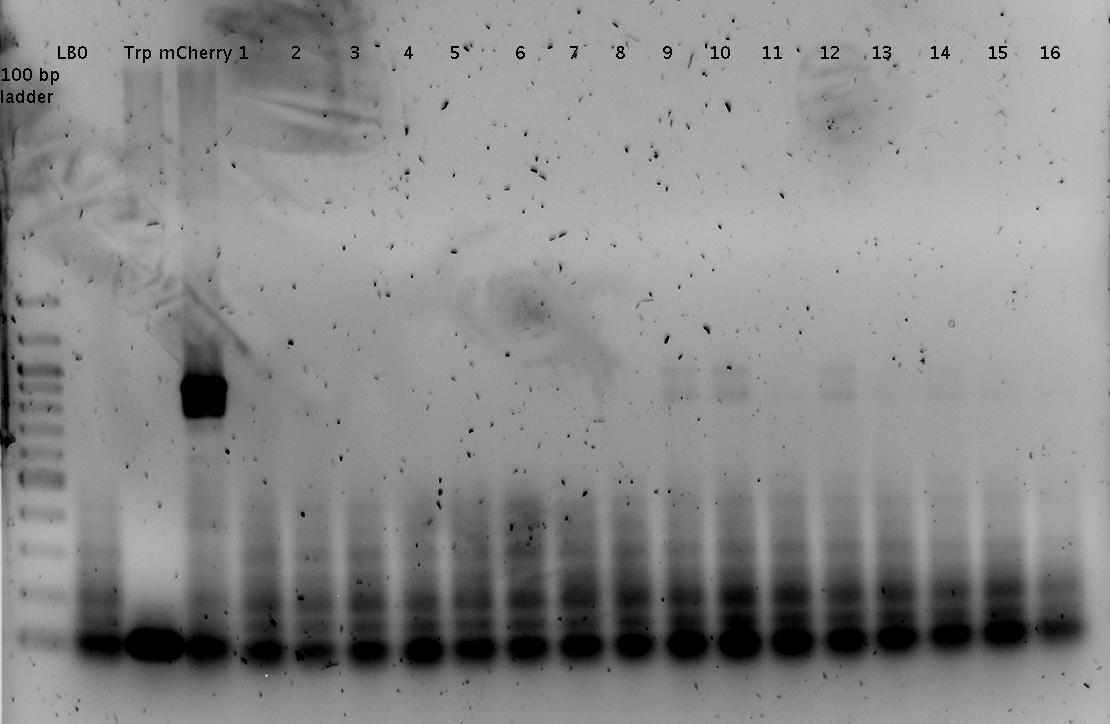
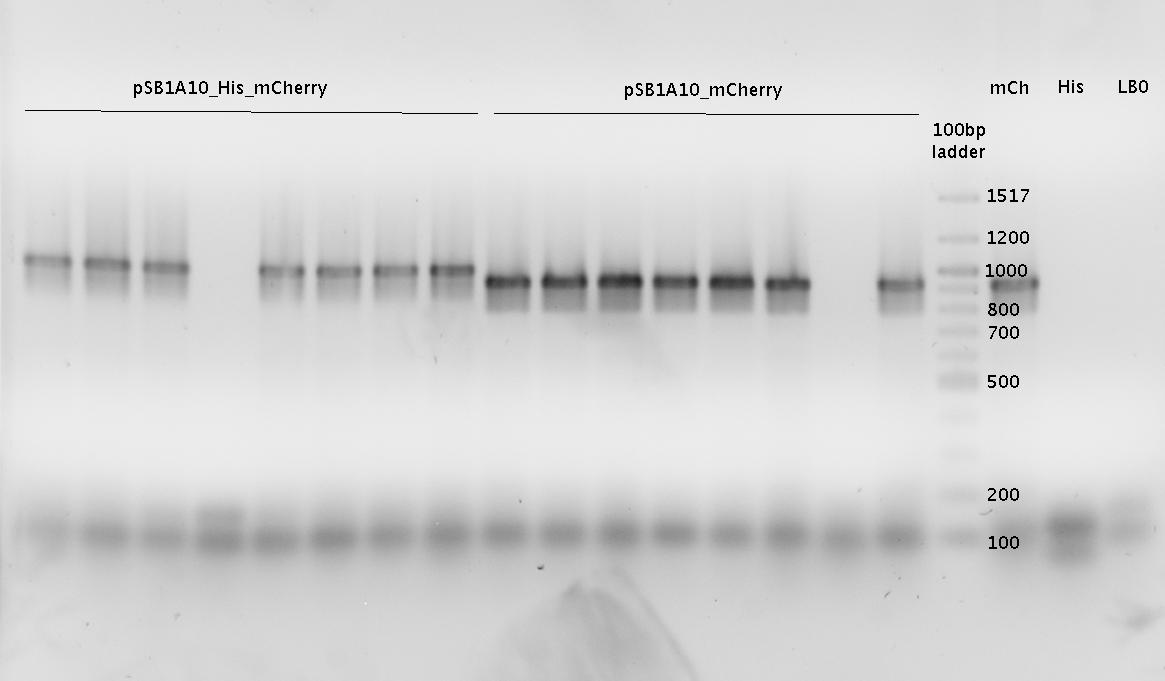
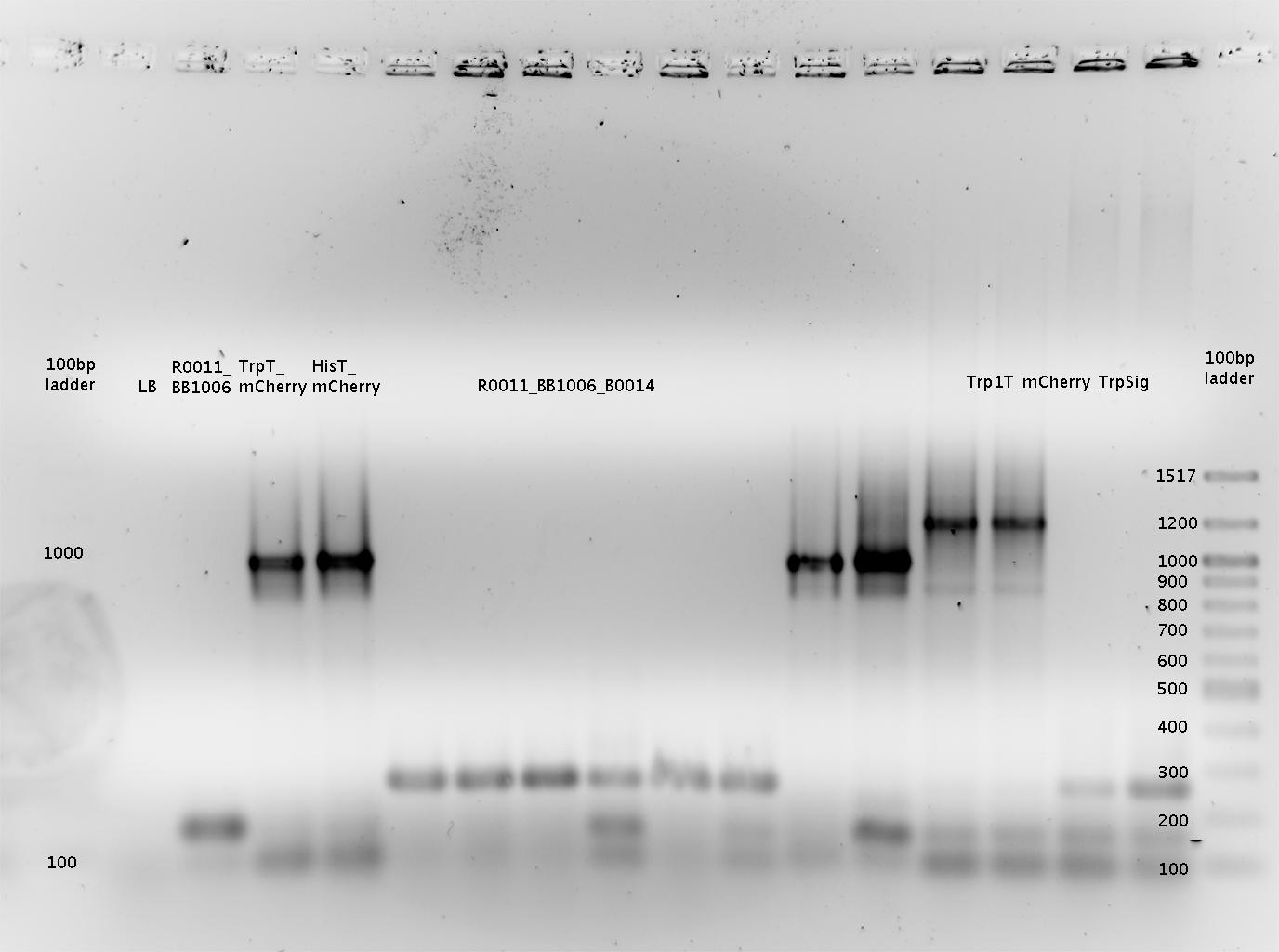
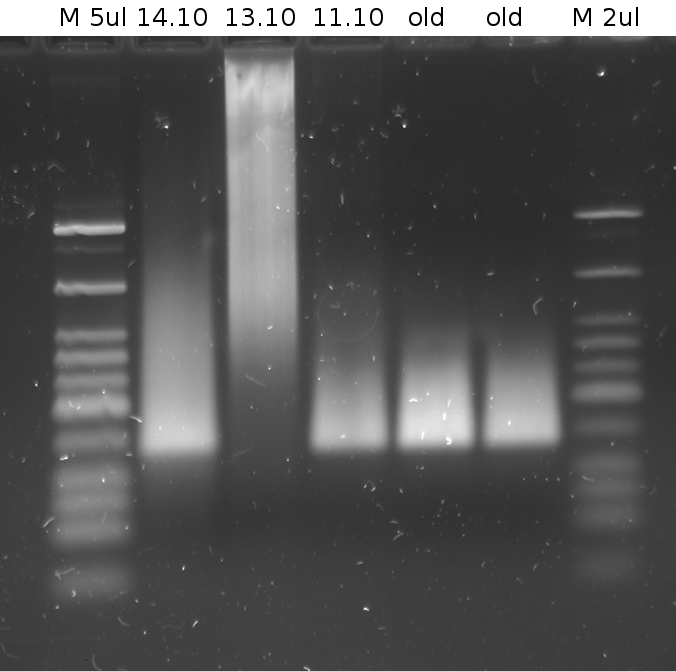
'''Gel of PCR products from 08.04.2010''' | '''Gel of PCR products from 08.04.2010''' | ||
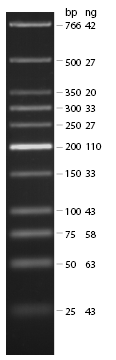
| - | *loaded: 10 | + | *loaded: 10 µL sample+2 µL 6x GLD, 4/2 µL LMW standard |
| - | * 110 V, | + | * 110 V, 90 min |
| - | *stained with Sybrgold, | + | *stained with Sybrgold, 20 min, 1:10.000 dilution in TAE |
| - | *Standard - Control - R0011_His - R0011_Trp - Standard(=low molecular weight (see [[ | + | *Standard - Control - R0011_His - R0011_Trp - Standard(=low molecular weight (see [[Team:TU_Munich/Lab#Molecular_Biology Lab_Protocols]])) |
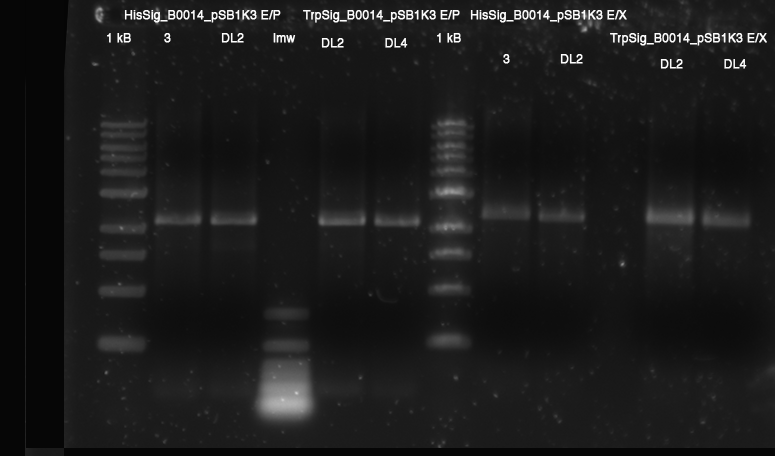
[[Image:TUM2010_100409.JPG]] | [[Image:TUM2010_100409.JPG]] | ||
| Line 628: | Line 203: | ||
===15.04.2010=== | ===15.04.2010=== | ||
Philipp & Flo | Philipp & Flo | ||
| - | <br><br>[http://web.e14.physik.tu-muenchen.de/igem/index.php/ | + | <br><br>[http://web.e14.physik.tu-muenchen.de/igem/index.php/Team:TU_Munich/Lab#Molecular_Biology PCR PCR] of B0014 and R0011 |
===16.04.2010=== | ===16.04.2010=== | ||
Philipp & Flo<br><br> | Philipp & Flo<br><br> | ||
| - | *'''Purification''' of PCR products from [[15.04.2010]] using [[ | + | *'''Purification''' of PCR products from [[15.04.2010]] using [[Team:TU_Munich/Lab#Molecular_Biology QIAquick_purification_Kit|QIA kit]] <br><br> |
*'''Concentrations''' measured with nanodrop:<br><br> | *'''Concentrations''' measured with nanodrop:<br><br> | ||
| Line 640: | Line 215: | ||
|- | |- | ||
| B0014 | | B0014 | ||
| - | | 2.5 ng/ | + | | 2.5 ng/µL<br> |
|- | |- | ||
| R0011<br> | | R0011<br> | ||
| - | | 27.5 ng/ | + | | 27.5 ng/µL<br> |
|} | |} | ||
<center><br> --> worked for R0011, not for B0014 <br> </center> | <center><br> --> worked for R0011, not for B0014 <br> </center> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|PCR]] '''of B0014<br> |
| - | **Purification with the [[ | + | **Purification with the [[Team:TU_Munich/Lab#Molecular_Biology ZYMO_RESEARCH_DNA_Clean.26Concentration_Kit|Zymo Kit]], Elution in 20 µL H2O |
| - | **Concentration measured with nanodrop, 17.5 ng/ | + | **Concentration measured with nanodrop, 17.5 ng/µL --> worked<br><br> |
*'''Digestions''' of:<br><br> | *'''Digestions''' of:<br><br> | ||
| Line 660: | Line 235: | ||
| EcoRI, PstI<br> | | EcoRI, PstI<br> | ||
|- | |- | ||
| - | | R0011 (from PCR [15042010], 27.5 ng/ | + | | R0011 (from PCR [15042010], 27.5 ng/µL<br> |
| SpeI<br> | | SpeI<br> | ||
|- | |- | ||
| Line 669: | Line 244: | ||
| XbaI<br> | | XbaI<br> | ||
|- | |- | ||
| - | | psB1K3 (with RFP insert, from HiWiPhilipp, 81 ng/ | + | | psB1K3 (with RFP insert, from HiWiPhilipp, 81 ng/µL) |
| EcoRI, PstI | | EcoRI, PstI | ||
|} | |} | ||
<br> | <br> | ||
| - | <center>5 | + | <center>5 µL template used for each setup. [[Team:TU_Munich/Lab#Molecular_Biology Restriction|protocol]] followed</center> |
<br> | <br> | ||
| Line 680: | Line 255: | ||
**2% Agarose in 1x TAE | **2% Agarose in 1x TAE | ||
**120 V, 90 min | **120 V, 90 min | ||
| - | **[[ | + | **[[Team:TU_Munich/Lab#Molecular_Biology stain|stained]] with SybrGold |
| - | **digestion, digestion, [[ | + | **digestion, digestion, [[Team:TU_Munich/Lab#Molecular_Biology standards|1 kb ladder]] |
***Digestion worked (partly). band at 2000 bp (backbone) cut<br><br> | ***Digestion worked (partly). band at 2000 bp (backbone) cut<br><br> | ||
[[Image:TUM2010_100416.png]]<br> | [[Image:TUM2010_100416.png]]<br> | ||
| Line 687: | Line 262: | ||
<br><br> | <br><br> | ||
*'''Purification of DNA from Gel''' | *'''Purification of DNA from Gel''' | ||
| - | **using the [[ | + | **using the [[Team:TU_Munich/Lab#Molecular_Biology ZYMO_RESEARCH_DNA_Clean.26Concentration_Kit|Zymo Kit]] |
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Ligation|Ligation]]''' of HisSig/TrpSig with R0011in 2 reactions<br><br> |
{| width="617" cellspacing="1" cellpadding="1" border="1" align="center" style="" | {| width="617" cellspacing="1" cellpadding="1" border="1" align="center" style="" | ||
| Line 700: | Line 275: | ||
|- | |- | ||
| HisSig<br> | | HisSig<br> | ||
| - | | 6 | + | | 6 µL<br> |
| - | | 7 ng/ | + | | 7 ng/µL<br> |
|- | |- | ||
| TrpSig<br> | | TrpSig<br> | ||
| - | | 6 | + | | 6 µL<br> |
| - | | 5 ng/ | + | | 5 ng/µL<br> |
|- | |- | ||
| R0011<br> | | R0011<br> | ||
| - | | 3 | + | | 3 µL<br> |
| - | | 6 ng/ | + | | 6 ng/µL<br> |
|} | |} | ||
| Line 731: | Line 306: | ||
{{:Team:TU Munich/Templates/ToggleBoxStart3}} | {{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
===19.04.2010=== | ===19.04.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|PCR]] '''of R0011-TrpSig and R0011-HisSig<br> |
| - | **Purification with the [[ | + | **Purification with the [[Team:TU_Munich/Lab#Molecular_Biology ZYMO_RESEARCH_DNA_Clean.26Concentration_Kit|Zymo Kit]], Elution in 30 uL H2O |
**Concentration measured with nanodrop: c(R0011-TrpSig)=20 ng/µL, c(R0011-HisSig)=12.5 ng/µl --> worked<br><br> | **Concentration measured with nanodrop: c(R0011-TrpSig)=20 ng/µL, c(R0011-HisSig)=12.5 ng/µl --> worked<br><br> | ||
| Line 738: | Line 313: | ||
**2% Agarose in 1x TAE | **2% Agarose in 1x TAE | ||
**110 V, 90 min | **110 V, 90 min | ||
| - | **[[ | + | **[[Team:TU_Munich/Lab#Molecular_Biology stain|stained]] with SybrGold 1:10000 20 min |
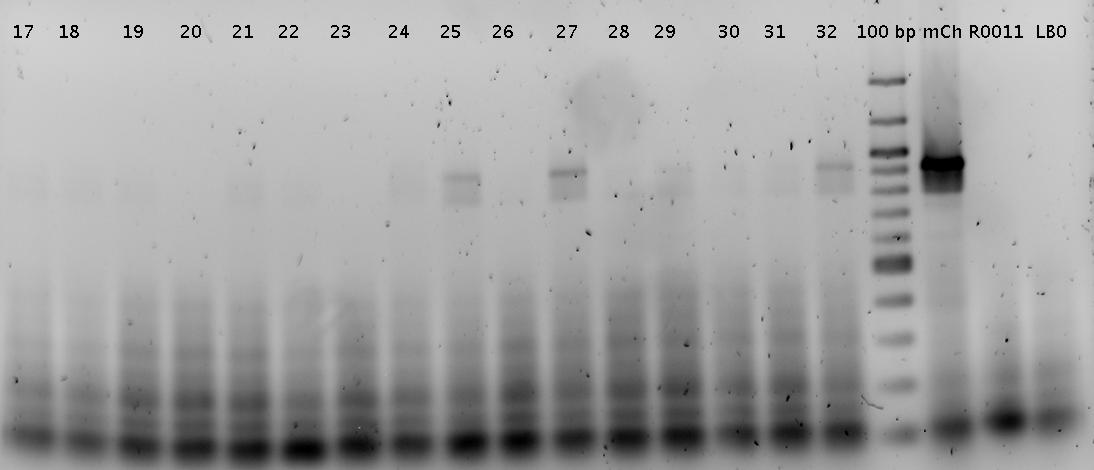
**pure R0011 PCR product used as control | **pure R0011 PCR product used as control | ||
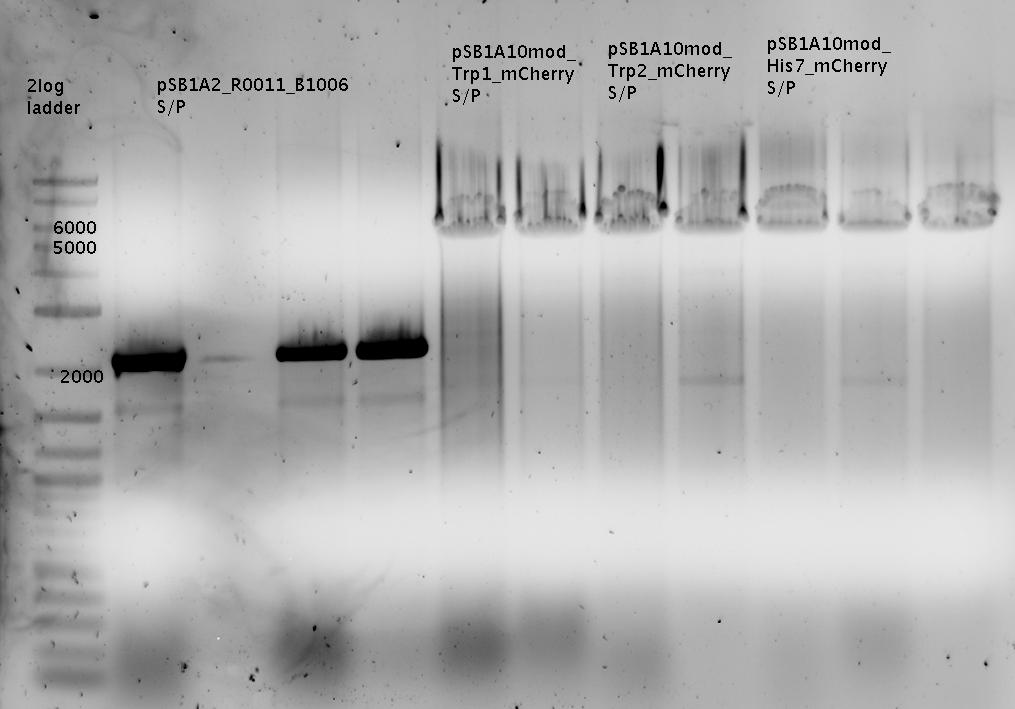
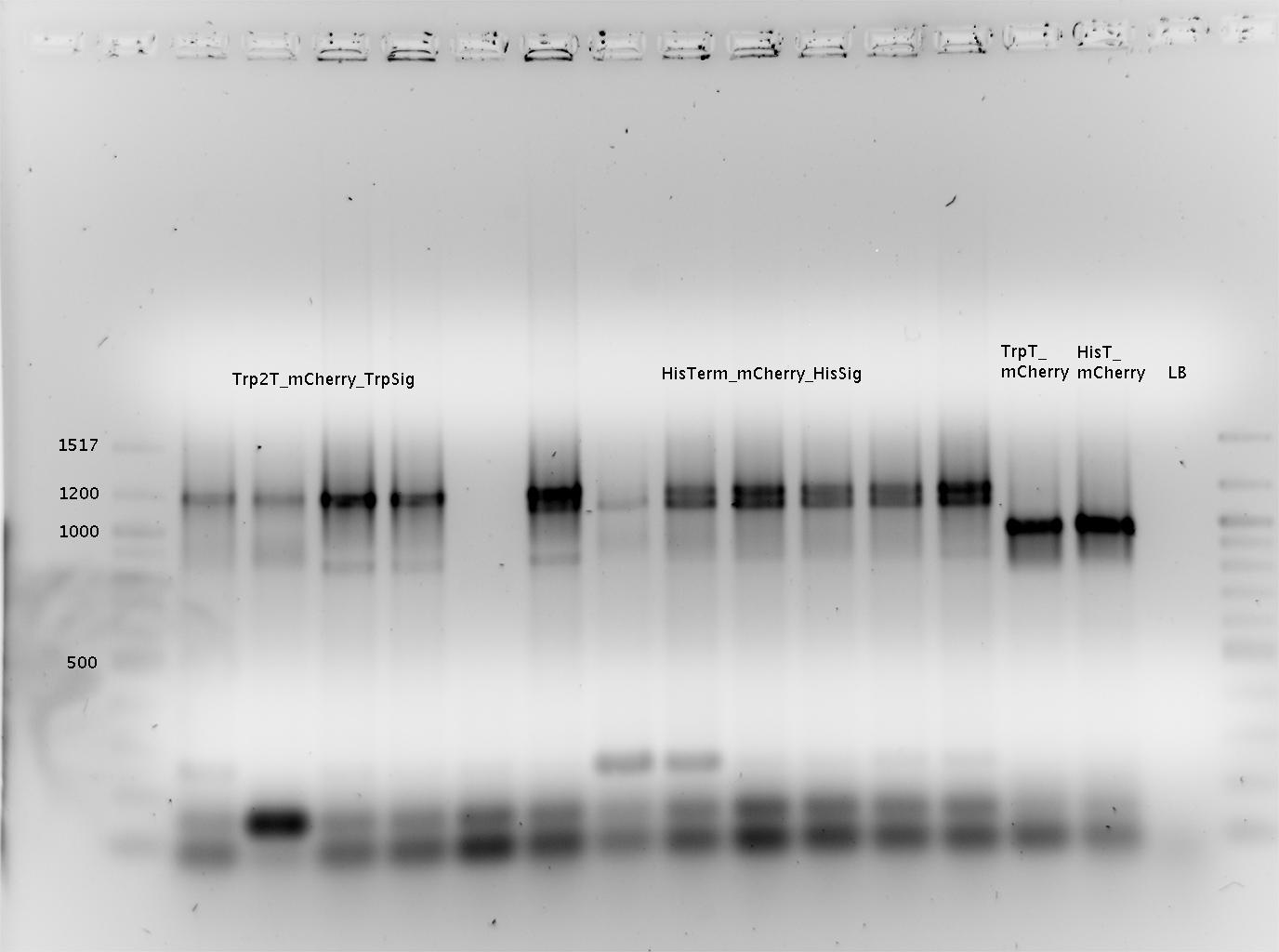
<br>[[Image:TUM2010_GEL_20100419beschriftet.png]] <br> | <br>[[Image:TUM2010_GEL_20100419beschriftet.png]] <br> | ||
| Line 762: | Line 337: | ||
<br> | <br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Ligation|Ligation]]''' of HisSig/TrpSig with R0011in 2 reactions<br><br> |
{| width="617" cellspacing="1" cellpadding="1" border="1" align="center" style="" | {| width="617" cellspacing="1" cellpadding="1" border="1" align="center" style="" | ||
| Line 771: | Line 346: | ||
|- | |- | ||
| pSB1K3<br> | | pSB1K3<br> | ||
| - | | 5 | + | | 5 µL<br> |
| 10 ng/µL (nanodrop)<br> | | 10 ng/µL (nanodrop)<br> | ||
|- | |- | ||
| B0014<br> | | B0014<br> | ||
| - | | 3 | + | | 3 µL<br> |
| 5 ng/µL approx.*<br> | | 5 ng/µL approx.*<br> | ||
| Line 787: | Line 362: | ||
**2% Agarose in 1x TAE | **2% Agarose in 1x TAE | ||
**130 V, 75 min | **130 V, 75 min | ||
| - | **[[ | + | **[[Team:TU_Munich/Lab#Molecular_Biology stain|stained]] with SybrGold 1:10000 60 min |
**pure R0011 PCR product used as control | **pure R0011 PCR product used as control | ||
| - | **Excision and purification of marked bands at | + | **Excision and purification of marked bands at 200 bp using QIA Kit, elution in 30 µl H2O<br> |
| - | [[Image:TUM2010_Gel100420marked.png | + | [[Image:TUM2010_Gel100420marked.png ]] |
<br><br><br> | <br><br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|PCR]] '''of excised and purified bands of R0011-TrpSig and R0011-HisSig<br> |
**complete samples (30 µl) used as templates | **complete samples (30 µl) used as templates | ||
| - | **Purification with the [[ | + | **Purification with the [[Team:TU_Munich/Lab#Molecular_Biology ZYMO_RESEARCH_DNA_Clean.26Concentration_Kit|Zymo Kit]], Elution in 30 uL H2O |
**Concentrations of PCR-products: 0.5-1 ng/µl --> Gel excision or PCR didn't work | **Concentrations of PCR-products: 0.5-1 ng/µl --> Gel excision or PCR didn't work | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Transformation|Transformation]] ''' (Woehlke-Lab) |
**8 µl of ligation product pSB1K3-B0014 to 50 µl XL-10 competent cells | **8 µl of ligation product pSB1K3-B0014 to 50 µl XL-10 competent cells | ||
**200 µl plated on a Kanamycin-containing Plate | **200 µl plated on a Kanamycin-containing Plate | ||
| Line 809: | Line 384: | ||
**2% Agarose in 1x TAE | **2% Agarose in 1x TAE | ||
**110 V, 90 min | **110 V, 90 min | ||
| - | **[[ | + | **[[Team:TU_Munich/Lab#Molecular_Biology stain|stained]] with SybrGold 1:10000 80 min |
**pure R0011 PCR product used as control | **pure R0011 PCR product used as control | ||
| - | **Excision and purification of marked bands at | + | **Excision and purification of marked bands at 200 bp using Zymo 5 Kit, elution in 20 µl H2O<br> |
[[Image:TUM2010_100421beschriftet.gif??]] | [[Image:TUM2010_100421beschriftet.gif??]] | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|PCR]] '''of excised and purified bands of R0011-TrpSig and R0011-HisSig<br> |
**complete samples (20 µl) used as templates | **complete samples (20 µl) used as templates | ||
| - | **Purification with the [[ | + | **Purification with the [[Team:TU_Munich/Lab#Molecular_Biology ZYMO_RESEARCH_DNA_Clean.26Concentration_Kit|Zymo Kit]], Elution in 25 uL H2O |
**Concentrations of PCR-products: | **Concentrations of PCR-products: | ||
*** R0011-TrpSig: 22.5 ng/µl | *** R0011-TrpSig: 22.5 ng/µl | ||
| Line 824: | Line 399: | ||
--> worked!!!!! | --> worked!!!!! | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]] ''' |
| - | ** | + | **7 Colonies picked and resuspended in 20 µl LB+Kana (each) |
**PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR, modified) | **PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR, modified) | ||
**15 µl of each sample mixed with 3 µl GLPn and loaded to Gel | **15 µl of each sample mixed with 3 µl GLPn and loaded to Gel | ||
| - | [[Image:TUM2010_100421colony.png | + | [[Image:TUM2010_100421colony.png]] |
<br><br> | <br><br> | ||
*Overnight cultures: | *Overnight cultures: | ||
| Line 840: | Line 415: | ||
**2% Agarose in 1x TAE | **2% Agarose in 1x TAE | ||
**110 V, 90 min | **110 V, 90 min | ||
| - | **[[ | + | **[[Team:TU_Munich/Lab#Molecular_Biology stain|stained]] with SybrGold 1:10000 30 min |
**pure R0011 PCR product used as control | **pure R0011 PCR product used as control | ||
**Excision and purification of marked bands at 200bp using Zymo 5 Kit, elution in 20 µl H2O<br> | **Excision and purification of marked bands at 200bp using Zymo 5 Kit, elution in 20 µl H2O<br> | ||
| - | [[Image:TUM2010_100422beschriftet.png | + | [[Image:TUM2010_100422beschriftet.png]] |
<br><br> | <br><br> | ||
| Line 851: | Line 426: | ||
===23.04.2010=== | ===23.04.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Restriction|Digestion]]''' of:<br><br> |
{| width="618" cellspacing="1" cellpadding="1" border="1" align="center" style="" | {| width="618" cellspacing="1" cellpadding="1" border="1" align="center" style="" | ||
| Line 883: | Line 458: | ||
**2% Agarose in 1x TAE (leftover from yesterday) | **2% Agarose in 1x TAE (leftover from yesterday) | ||
**140 V, 90 min | **140 V, 90 min | ||
| - | **[[ | + | **[[Team:TU_Munich/Lab#Molecular_Biology stain|stained]] with SybrGold 40 min |
| - | **4 µl [[ | + | **4 µl [[Team:TU_Munich/Lab#Molecular_Biology standards|1 kb ladder]], 10 µl purified digestion + 2 µl GLPn, 10 µl purified digestion + 2 µl GLPn |
***Digestion worked (partly). band at 2400 bp cut out<br><br> | ***Digestion worked (partly). band at 2400 bp cut out<br><br> | ||
| - | [[Image:TUM2010_100423beschriftet.png | + | [[Image:TUM2010_100423beschriftet.png]]<br> |
<br><br> | <br><br> | ||
*'''Purification of DNA from Gel''' | *'''Purification of DNA from Gel''' | ||
| - | **using the [[ | + | **using the [[Team:TU_Munich/Lab#Molecular_Biology ZYMO_RESEARCH_Gel_DNA_Recovery_Kit|Zymo Kit]] |
**elution in 25 µl H2O | **elution in 25 µl H2O | ||
* A260/A230 and A260/A280 values were strange (see labbook) | * A260/A230 and A260/A280 values were strange (see labbook) | ||
| Line 900: | Line 475: | ||
===26.04.2010=== | ===26.04.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Digestion|Digestion]]''' of pSB1K3-B0014 with EcorI and XbaI |
** 10 µl template (No1, 50 ng/µl) | ** 10 µl template (No1, 50 ng/µl) | ||
** 5 µl BSA, 5 µl Buffer NEB#4 | ** 5 µl BSA, 5 µl Buffer NEB#4 | ||
| Line 909: | Line 484: | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Ligation|Ligation]]''' of Signals and PSB1K3-B0014 |
**3 µl of each sample, end volume 20 µl | **3 µl of each sample, end volume 20 µl | ||
<br> | <br> | ||
*Preparation of Measurement Plasmid from Folder, Transformation | *Preparation of Measurement Plasmid from Folder, Transformation | ||
| - | **Plate 1022, Spots 1E, 1G, 2A: pSB1A10 with different Inserts, all inserts are Zinc-finger constructs with about 1 | + | **Plate 1022, Spots 1E, 1G, 2A: pSB1A10 with different Inserts, all inserts are Zinc-finger constructs with about 1.6 kb |
<br> | <br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Transformation|Transformation]] ''' of XL10 with Ligation Products (8 µl each) and pSB1A10 (2 µl each) |
<br> | <br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Preparation of BioBricks from distribution 2008|Preparation]]''' of Measurement Plasmid from Folder, Transformation |
| - | **Plate 1022, Spots 1E, 1G, 2A: pSB1A10 with different Inserts, all inserts are Zinc-finger constructs with about 1 | + | **Plate 1022, Spots 1E, 1G, 2A: pSB1A10 with different Inserts, all inserts are Zinc-finger constructs with about 1.6 kb |
<br> | <br> | ||
*growing over night cultures of remaining PSB1K3-B0014-transformed cells | *growing over night cultures of remaining PSB1K3-B0014-transformed cells | ||
| Line 926: | Line 501: | ||
*Plenty of cultures on both HisSig and TrpSig Ligation plates, but nothing on pSB1A10 plates! --> repeat DNA extraction, ask Prof. Simmel for new Distribution | *Plenty of cultures on both HisSig and TrpSig Ligation plates, but nothing on pSB1A10 plates! --> repeat DNA extraction, ask Prof. Simmel for new Distribution | ||
<br> | <br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]]''' |
| - | ** | + | **7 Colonies picked and resuspended in 20 µl LB+Kana (each) |
**PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR, modified) | **PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR, modified) | ||
**10 µl of each sample mixed with 2 µl GLPn and loaded to Gel | **10 µl of each sample mixed with 2 µl GLPn and loaded to Gel | ||
| - | [[Image:TUM2010_100427beschriftet.png | + | [[Image:TUM2010_100427beschriftet.png]] |
Many colonies with pSB1K3-B0014, not one with pSB1K3-Sig-B0014 | Many colonies with pSB1K3-B0014, not one with pSB1K3-Sig-B0014 | ||
| Line 947: | Line 522: | ||
--> Better results for 600 µl cultures without centrifuging!!! | --> Better results for 600 µl cultures without centrifuging!!! | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Preparation of BioBricks from distribution 2008|Preparation]]''' of Measurement Plasmid from Folder, Transformation |
| - | **Plate 1022, Spots 1E, 1F, 1G, 1H, 2A: pSB1A10 with different Inserts, all inserts are Zinc-finger constructs with about 1 | + | **Plate 1022, Spots 1E, 1F, 1G, 1H, 2A: pSB1A10 with different Inserts, all inserts are Zinc-finger constructs with about 1.6 kb |
<br> | <br> | ||
| - | |||
===28.04.2010=== | ===28.04.2010=== | ||
| Line 958: | Line 532: | ||
<br> | <br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]]''' |
**PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR, modified) | **PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR, modified) | ||
**15 µl of each sample mixed with 3 µl GLPn and loaded to Gel | **15 µl of each sample mixed with 3 µl GLPn and loaded to Gel | ||
| Line 966: | Line 540: | ||
<br> | <br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Digestion|Digestion]]''' of pSB1K3-B0014 with EcorI and XbaI |
** 10 µl template (No1, 50 ng/µl) | ** 10 µl template (No1, 50 ng/µl) | ||
** 5 µl BSA, 5 µl Buffer NEB#4 | ** 5 µl BSA, 5 µl Buffer NEB#4 | ||
| Line 973: | Line 547: | ||
** 1.5 h @ 37°C | ** 1.5 h @ 37°C | ||
** heat inactivation 5min @60°C | ** heat inactivation 5min @60°C | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Dephosphorylation|Dephosphorylation]]''' of restricted vector |
**Purification with Zymo5 Kit, elution in 20 µl H2O | **Purification with Zymo5 Kit, elution in 20 µl H2O | ||
**loaded on gel (with 4 µl GLPn) (Gel shown above) | **loaded on gel (with 4 µl GLPn) (Gel shown above) | ||
| Line 984: | Line 558: | ||
===29.04.2010=== | ===29.04.2010=== | ||
<br> | <br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Digestion|Digestion]]''' of pSB1K3-B0014 with EcorI and XbaI |
** 10 µl template (NoIV, 108 ng/µl) | ** 10 µl template (NoIV, 108 ng/µl) | ||
** 5 µl BSA, 5 µl Buffer NEB#4 | ** 5 µl BSA, 5 µl Buffer NEB#4 | ||
| Line 991: | Line 565: | ||
** 1.5 h @ 37°C | ** 1.5 h @ 37°C | ||
** heat inactivation 5min @60°C | ** heat inactivation 5min @60°C | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Dephosphorylation|Dephosphorylation]]''' of restricted vector |
**Purification with Zymo5 Kit, elution in 20 µl H2O | **Purification with Zymo5 Kit, elution in 20 µl H2O | ||
**loaded on gel (with 4 µl GLPn) | **loaded on gel (with 4 µl GLPn) | ||
| Line 999: | Line 573: | ||
** | ** | ||
<br> | <br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Digestion|Digestion]]''' of R0011 with SpeI |
** 10 µl template (R0011, X ng/µl) | ** 10 µl template (R0011, X ng/µl) | ||
** 5 µl BSA, 5 µl Buffer NEB#4 | ** 5 µl BSA, 5 µl Buffer NEB#4 | ||
| Line 1,005: | Line 579: | ||
** 29 µl H2O | ** 29 µl H2O | ||
** 1.5 h @ 37°C | ** 1.5 h @ 37°C | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Ligation|Ligation]]''' |
** 5 µl R0011 (S-digested) with 12 µl TrpSig or HisSig, respectively (X-digested) | ** 5 µl R0011 (S-digested) with 12 µl TrpSig or HisSig, respectively (X-digested) | ||
** complete ligation (20 µl) loaded on Gel (with 4 µl GLPn) | ** complete ligation (20 µl) loaded on Gel (with 4 µl GLPn) | ||
| - | [[Image:TUM2010_100429beschriftet.png | + | [[Image:TUM2010_100429beschriftet.png]] |
** Gel excision with Zymo Kit, eluted in 42 µl H2O | ** Gel excision with Zymo Kit, eluted in 42 µl H2O | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Transformation|Transformation]]''' |
**50 µl XL-10 transformed with 2 µl of pSB1A10 prepared from IGem 2009 Distribution (13 µl left in pink Box @-20°C) | **50 µl XL-10 transformed with 2 µl of pSB1A10 prepared from IGem 2009 Distribution (13 µl left in pink Box @-20°C) | ||
* | * | ||
| Line 1,026: | Line 600: | ||
{{:Team:TU Munich/Templates/ToggleBoxStart3}} | {{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
===04.05.2010=== | ===04.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Digestion|Digestion]]''' of pSB1K3-B0014 with EcorI and XbaI |
** 10 µl template (NoIII, 103 ng/µl) | ** 10 µl template (NoIII, 103 ng/µl) | ||
** 5 µl BSA, 5 µl Buffer NEB#4 | ** 5 µl BSA, 5 µl Buffer NEB#4 | ||
| Line 1,034: | Line 608: | ||
**Purification with Zymo5 Kit, elution in 15 µl H2O | **Purification with Zymo5 Kit, elution in 15 µl H2O | ||
**loaded on gel (with 3 µl GLPn) | **loaded on gel (with 3 µl GLPn) | ||
| - | [[Image:TUM2010_100504beschriftet.png | + | [[Image:TUM2010_100504beschriftet.png]] |
*Gel excision with Zymo Kit | *Gel excision with Zymo Kit | ||
** | ** | ||
** | ** | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Digestion|Digestion]]''' of R0011-HisSig and R0011-TrpSig with EcorI and SpeI |
** 10 µl template (PCR-product) | ** 10 µl template (PCR-product) | ||
** 5 µl BSA, 5 µl Buffer NEB#4 | ** 5 µl BSA, 5 µl Buffer NEB#4 | ||
| Line 1,046: | Line 620: | ||
**Purification with Zymo5 Kit, elution in 20 µl H2O | **Purification with Zymo5 Kit, elution in 20 µl H2O | ||
<br> | <br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Ligation|Ligation]]''' |
** 4 µl R0011-Signal (E/S-digested) with 4 µl pSB1K3-B0014 (E/X-digested) | ** 4 µl R0011-Signal (E/S-digested) with 4 µl pSB1K3-B0014 (E/X-digested) | ||
**15 min @ RT, 20 min heat inactivation @ 65°C | **15 min @ RT, 20 min heat inactivation @ 65°C | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Transformation|Transformation]]''' |
**50 µl XL-10 transformed with 10 µl of Ligation mix | **50 µl XL-10 transformed with 10 µl of Ligation mix | ||
<br> | <br> | ||
| Line 1,061: | Line 635: | ||
--> each in 600 µl LB+Carbenicillin (=Ampicillin) @37°C | --> each in 600 µl LB+Carbenicillin (=Ampicillin) @37°C | ||
===05.05.2010=== | ===05.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Miniprep|Miniprep]]''' of pSB1A10; 4 samples (1, 2; 3a; 3b) |
**eluted in 50 µl H2O each | **eluted in 50 µl H2O each | ||
**Concentrations: | **Concentrations: | ||
| - | *** c1=37 | + | *** c1=37.5 ng/µl |
| - | *** c2=56 | + | *** c2=56.5 ng/µl |
| - | *** c3a=46 | + | *** c3a=46.5 ng/µl |
*** c3b=30 ng/µl | *** c3b=30 ng/µl | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Digestion|Digestion]]''' of pSB1A10 with EcorI and PstI, 4 samples (1, 2; 3a; 3b) |
** 15 µl template | ** 15 µl template | ||
** 5 µl BSA, 5 µl Buffer NEB#3 | ** 5 µl BSA, 5 µl Buffer NEB#3 | ||
| Line 1,078: | Line 652: | ||
**Purification with Zymo5 Kit, elution in 15 µl H2O | **Purification with Zymo5 Kit, elution in 15 µl H2O | ||
**loaded on gel (with 3 µl GLPn) | **loaded on gel (with 3 µl GLPn) | ||
| - | [[Image:TUM2010_100505beschriftet.png | + | [[Image:TUM2010_100505beschriftet.png]] |
<br> | <br> | ||
| - | Insert @ 1 kb as expected, but vector @ | + | Insert @ 1 kb as expected, but vector @ 2 kb and not @ 5 kb as expected!!!! |
--> Wrong Plasmid! Comparison to the [http://partsregistry.org/cgi/assembly/plate_egel.cgi?id=615 Gel in the registry] shows: The Distribution contains the wrong plasmid! | --> Wrong Plasmid! Comparison to the [http://partsregistry.org/cgi/assembly/plate_egel.cgi?id=615 Gel in the registry] shows: The Distribution contains the wrong plasmid! | ||
<br> | <br> | ||
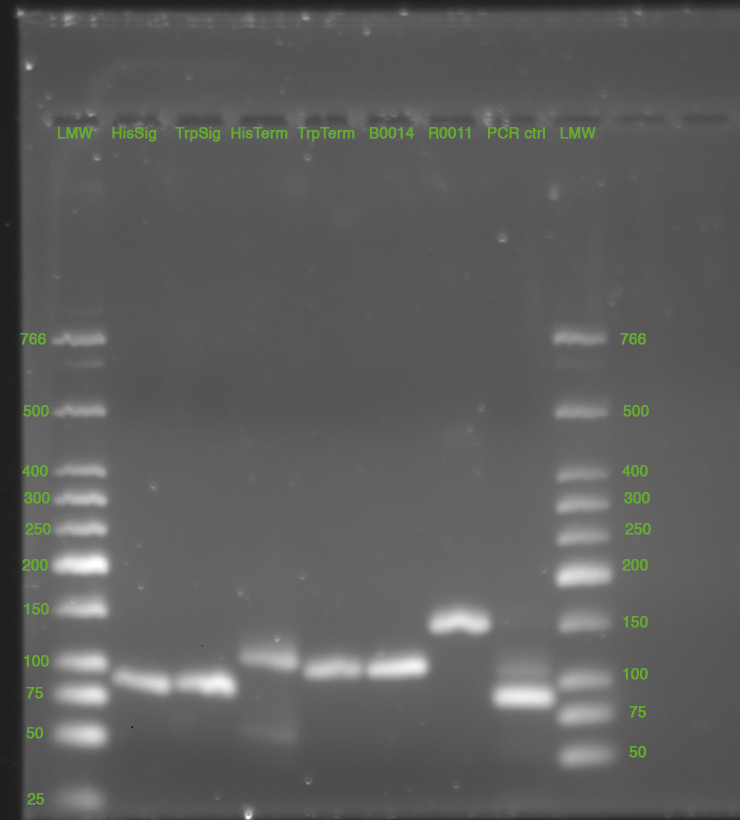
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Digestion|Digestion]]''' of HisTerm and TrpTerm with EcorI and PstI |
** 5 µl template | ** 5 µl template | ||
** 5 µl BSA, 5 µl Buffer NEB#3 | ** 5 µl BSA, 5 µl Buffer NEB#3 | ||
| Line 1,091: | Line 665: | ||
<br><br> | <br><br> | ||
*Clones picked: 7 from each Plate (pSB1K3-R0011-TrpSig-Boo14 and pSB1K3-R0011-HisSig-Boo14) | *Clones picked: 7 from each Plate (pSB1K3-R0011-TrpSig-Boo14 and pSB1K3-R0011-HisSig-Boo14) | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]]''' |
**PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR) | **PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR) | ||
**15 µl of each sample mixed with 3 µl GLPn and loaded to Gel | **15 µl of each sample mixed with 3 µl GLPn and loaded to Gel | ||
| Line 1,101: | Line 675: | ||
===06.05.2010=== | ===06.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Digestion|Digestion]]''' of pSB1K3 with EcorI and XbaI |
** 20 µl template (sample III, 103 ng/µl) | ** 20 µl template (sample III, 103 ng/µl) | ||
** 5 µl BSA, 5 µl Buffer NEB#3 | ** 5 µl BSA, 5 µl Buffer NEB#3 | ||
| Line 1,109: | Line 683: | ||
** heat inactivation 5min @60°C | ** heat inactivation 5min @60°C | ||
**loaded on gel (with 10 µl GLPn) in 4 lanes | **loaded on gel (with 10 µl GLPn) in 4 lanes | ||
| - | [[Image:TUM2010_100506beschriftet.png | + | [[Image:TUM2010_100506beschriftet.png]] |
*Gel excision with Zymo Kit (lanes 1&2) and with Qiaquick Kit (lanes 3&4) | *Gel excision with Zymo Kit (lanes 1&2) and with Qiaquick Kit (lanes 3&4) | ||
** c1=4.5 ng/µl | ** c1=4.5 ng/µl | ||
| Line 1,117: | Line 691: | ||
* A260/A230 and A260/A280 values were strange (see labbook) | * A260/A230 and A260/A280 values were strange (see labbook) | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Ligation|Ligation]]''' |
** 4 µl R0011-Signal (E/S-digested) with 10 µl pSB1K3-B0014 (E/X-digested, from [[23.04.2010|23.04.]]) | ** 4 µl R0011-Signal (E/S-digested) with 10 µl pSB1K3-B0014 (E/X-digested, from [[23.04.2010|23.04.]]) | ||
**15 min @ RT, 20 min heat inactivation @ 65°C | **15 min @ RT, 20 min heat inactivation @ 65°C | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Transformation|Transformation]]''' |
**50 µl XL-10 transformed with 7 µl of Ligation mix | **50 µl XL-10 transformed with 7 µl of Ligation mix | ||
<br> | <br> | ||
| Line 1,130: | Line 704: | ||
*Clones picked: 7 from each Plate (pSB1K3-R0011-TrpSig-Boo14 and pSB1K3-R0011-HisSig-Boo14) | *Clones picked: 7 from each Plate (pSB1K3-R0011-TrpSig-Boo14 and pSB1K3-R0011-HisSig-Boo14) | ||
<br> | <br> | ||
| - | |||
---Too damn stupid to do a PCR!!!---<br> <br> | ---Too damn stupid to do a PCR!!!---<br> <br> | ||
| - | |||
<br> | <br> | ||
* replated picked clones on new plates, incubated at RT | * replated picked clones on new plates, incubated at RT | ||
| Line 1,139: | Line 711: | ||
{{:Team:TU Munich/Templates/ToggleBoxStart1}}Week06{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/RedBox | text=''in vivo'' constructs }} | {{:Team:TU Munich/Templates/ToggleBoxStart1}}Week06{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/RedBox | text=''in vivo'' constructs }} | ||
{{:Team:TU Munich/Templates/ToggleBoxStart3}} | {{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
| + | |||
===10.05.2010=== | ===10.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]]''' of picked clones from [[07.05.2010|Fr 07.05.2010]] |
**PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR, modified) | **PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR, modified) | ||
**15 µl of each sample mixed with 3 µl GLPn and loaded to Gel | **15 µl of each sample mixed with 3 µl GLPn and loaded to Gel | ||
| Line 1,194: | Line 767: | ||
===11.05.2010=== | ===11.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Ligation|Ligation]]''' |
** 4 µl Signal (E/S-digested; from ) with 5 µl pSB1K3-B0014 (E/X-digested; from) | ** 4 µl Signal (E/S-digested; from ) with 5 µl pSB1K3-B0014 (E/X-digested; from) | ||
**15 min @ RT | **15 min @ RT | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Transformation|Transformation]]''' |
**50 µl XL-10 transformed with 7 µl of Ligation mix | **50 µl XL-10 transformed with 7 µl of Ligation mix | ||
<br> | <br> | ||
===12.05.2010=== | ===12.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]]''' of picked clones from [[11.05.2010|Tu 12.05.2010]] |
**PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR, modified) | **PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR, modified) | ||
**15 µl of each sample mixed with 3 µl GLPn and loaded to Gel | **15 µl of each sample mixed with 3 µl GLPn and loaded to Gel | ||
| Line 1,260: | Line 833: | ||
Prefix: 20 bp/29 bp after PCR; Suffix: 21 bp/30 bp after PCR; X-S-scar: 6 bp | Prefix: 20 bp/29 bp after PCR; Suffix: 21 bp/30 bp after PCR; X-S-scar: 6 bp | ||
===14.05.2010=== | ===14.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Digestion|Digestion]]''' of pSB1K3 with EcorI and XbaI |
** 10 µl template (sample III, 103 ng/µl) | ** 10 µl template (sample III, 103 ng/µl) | ||
** 2 µl BSA, 2 µl Buffer NEB#4 | ** 2 µl BSA, 2 µl Buffer NEB#4 | ||
| Line 1,267: | Line 840: | ||
** 1 h @ 37°C | ** 1 h @ 37°C | ||
**loaded on gel (with 4 µl GLPn) in 1 lane | **loaded on gel (with 4 µl GLPn) in 1 lane | ||
| - | [[Image:TUM2010_100514beschriftet.png | + | [[Image:TUM2010_100514beschriftet.png]] |
*Gel excision with Zymo Kit | *Gel excision with Zymo Kit | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Digestion|Digestion]]''' of HisSig and TrpSig with EcorI and SpeI |
** 10 µl template ("1:100") | ** 10 µl template ("1:100") | ||
** 2 µl BSA, 2 µl Buffer NEB#3 | ** 2 µl BSA, 2 µl Buffer NEB#3 | ||
| Line 1,278: | Line 851: | ||
** 4 µl H2O | ** 4 µl H2O | ||
** 1.5 h @ 37°C | ** 1.5 h @ 37°C | ||
| - | ** Purification with [[ | + | ** Purification with [[Team:TU_Munich/Lab#Molecular_Biology ZYMO RESEARCH DNA Clean&Concentration Kit|Zymo 5 ]] |
** or heat inactivated (20 min @ 80°C) | ** or heat inactivated (20 min @ 80°C) | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Ligation|Ligation]]''' |
** | ** | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Transformation|Transformation]]''' |
**50 µl XL-10 transformed with 10 µl of Ligation mix | **50 µl XL-10 transformed with 10 µl of Ligation mix | ||
**50 µl untransformed cells plated on Kana-plate as control | **50 µl untransformed cells plated on Kana-plate as control | ||
| Line 1,298: | Line 871: | ||
<br><br> | <br><br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Transformation|Transformation]]''' |
**50 µl DH5a transformed with 10 µl of Friday's Ligation mix | **50 µl DH5a transformed with 10 µl of Friday's Ligation mix | ||
**plated on Kana-Plates; Overnight @ 37°C | **plated on Kana-Plates; Overnight @ 37°C | ||
| Line 1,305: | Line 878: | ||
*DNA Isolation from BioBrick Distribution 2010 | *DNA Isolation from BioBrick Distribution 2010 | ||
** 10 µl H2O added to Well 1A of plate 1 containing pSB1A10 with RFP-insert | ** 10 µl H2O added to Well 1A of plate 1 containing pSB1A10 with RFP-insert | ||
| - | ** 2 µl used for [[ | + | ** 2 µl used for [[Team:TU_Munich/Lab#Molecular_Biology Transformation|Transformation]] of 50 µl DH5a-cells |
** plated on Carbenicillin (=Amp-analogon)-plates, Overnight @ 37°C | ** plated on Carbenicillin (=Amp-analogon)-plates, Overnight @ 37°C | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Preparation of Gels|Polyacrylamide Gel]]''' prepared for tomorrow |
** 1 big denaturing Gel with 20 pockets | ** 1 big denaturing Gel with 20 pockets | ||
===18.05.2010=== | ===18.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]]''' of picked clones |
**PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR) | **PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR) | ||
**10 µl of each sample mixed with 10 µl Formamide loading buffer and loaded to Polyacrylamide Gel | **10 µl of each sample mixed with 10 µl Formamide loading buffer and loaded to Polyacrylamide Gel | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Running of Gels|Polyacrylamide Gel]]''' |
Samples: | Samples: | ||
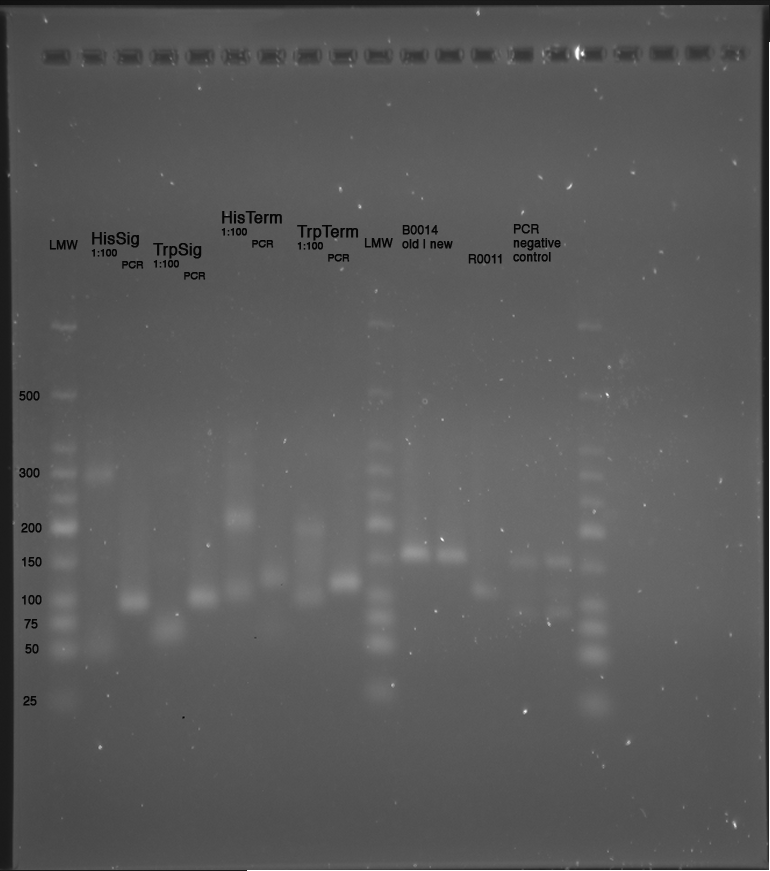
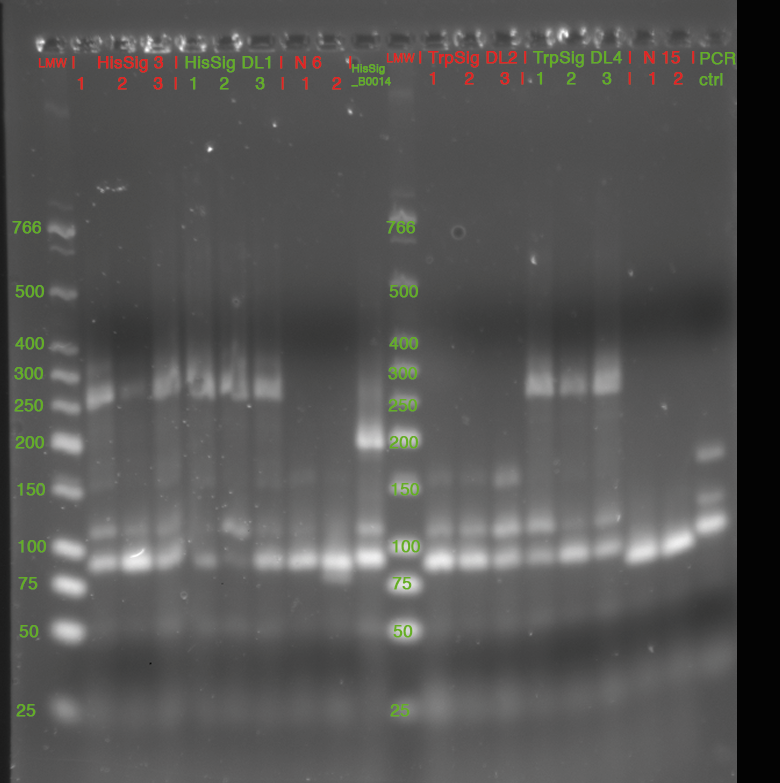
LMW|R0011|HisSig|TrpSig|HisSig E/S-Dig|TrpSig E/S-Dig|B0014|LMW2|Colony PCR His1|His2|Trp1|Trp2|control|HisTerm|TrpTerm|HisTerm E/P-Dig|TrpTerm E/P-Dig | LMW|R0011|HisSig|TrpSig|HisSig E/S-Dig|TrpSig E/S-Dig|B0014|LMW2|Colony PCR His1|His2|Trp1|Trp2|control|HisTerm|TrpTerm|HisTerm E/P-Dig|TrpTerm E/P-Dig | ||
| Line 1,335: | Line 908: | ||
(*R0011 and B0014 look normal | (*R0011 and B0014 look normal | ||
| - | *ColonyPCR: bands that look like B0014 in all clones (and in control | + | *ColonyPCR: bands that look like B0014 in all clones (and in control) --> Religation? |
*Signals at the wrong size: should be about 75 bp, look like 200 bp!!! | *Signals at the wrong size: should be about 75 bp, look like 200 bp!!! | ||
*terminators completely strange: should be around 100 bp! | *terminators completely strange: should be around 100 bp! | ||
| Line 1,345: | Line 918: | ||
*5 µl of each samples mixed with 5 µl formamide loading dye and loaded to gel(except Ladder and colonyPCR) | *5 µl of each samples mixed with 5 µl formamide loading dye and loaded to gel(except Ladder and colonyPCR) | ||
**LMW: 3 µl LMW (Korbinian) + 3 µl Formamide loading Dye | **LMW: 3 µl LMW (Korbinian) + 3 µl Formamide loading Dye | ||
| - | ** | + | **colony PCR: from Tuesday, 8 µl sample with 8 µl Formamide loading Dye |
*stained in SybrSafe 20 min | *stained in SybrSafe 20 min | ||
[[Image:TUM2010_100519paabeschriftet.png]]<br> | [[Image:TUM2010_100519paabeschriftet.png]]<br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]]''' |
** 4 colonies picked from each Plate (Ligations from yesterday; Signal-B0014) | ** 4 colonies picked from each Plate (Ligations from yesterday; Signal-B0014) | ||
**15 µl of each Sample mixed with 3 µl GLPn and loaded to Gel: | **15 µl of each Sample mixed with 3 µl GLPn and loaded to Gel: | ||
** 3% Agarose in 1x TBE, 130 V | ** 3% Agarose in 1x TBE, 130 V | ||
[[Image:TUM2010_100519beschriftet.png600px]] | [[Image:TUM2010_100519beschriftet.png600px]] | ||
| + | |||
===20.05.2010=== | ===20.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Digestion|Digestion]]''' |
** template | ** template | ||
** 2 µl BSA | ** 2 µl BSA | ||
| Line 1,396: | Line 970: | ||
**digested inserts heat inactivated (20 min @ 80°C) | **digested inserts heat inactivated (20 min @ 80°C) | ||
**digested plasmids loaded on gel (with 4 µl GLPn) in 1 lane | **digested plasmids loaded on gel (with 4 µl GLPn) in 1 lane | ||
| - | [[Image:TUM2010_100520beschriftet.png | + | [[Image:TUM2010_100520beschriftet.png]] |
*Gel excision with Zymo Kit | *Gel excision with Zymo Kit | ||
**c(pSB1A10, I)=4.5 ng/µl | **c(pSB1A10, I)=4.5 ng/µl | ||
| Line 1,408: | Line 982: | ||
[[Image:TUM2010_100520bbeschriftet.png]] | [[Image:TUM2010_100520bbeschriftet.png]] | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Ligation|Ligation]]''' |
** templates | ** templates | ||
**2 µl T4-buffer 10x | **2 µl T4-buffer 10x | ||
| Line 1,438: | Line 1,012: | ||
|} | |} | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Transformation|Transformation]]''' |
** 50 µl DH5a transformed with 10 µl of Ligation mix | ** 50 µl DH5a transformed with 10 µl of Ligation mix | ||
** 50 µl DH5a transformed with 2 µl of pSB1K3_B0014 | ** 50 µl DH5a transformed with 2 µl of pSB1K3_B0014 | ||
| Line 1,444: | Line 1,018: | ||
** 50 µl DH5a transformed with 2 µl of pSB1A10_RFP | ** 50 µl DH5a transformed with 2 µl of pSB1A10_RFP | ||
===21.05.2010=== | ===21.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]]''' |
** 4 colonies picked from each Plate (pSB1K3_HisSig_B0014, pSB1K3_TrpSig_B0014, pSB1K3_HisSig_B0014 double ligation, pSB1K3_TrpSig_B0014 double ligation) | ** 4 colonies picked from each Plate (pSB1K3_HisSig_B0014, pSB1K3_TrpSig_B0014, pSB1K3_HisSig_B0014 double ligation, pSB1K3_TrpSig_B0014 double ligation) | ||
** each clone resuspended in 20 µl LB0, 3 µl used as template for PCR | ** each clone resuspended in 20 µl LB0, 3 µl used as template for PCR | ||
** 15 µl of each Sample mixed with 3 µl GLPn and loaded to Gel: | ** 15 µl of each Sample mixed with 3 µl GLPn and loaded to Gel: | ||
** 3% Agarose in 1x TBE, 130 V | ** 3% Agarose in 1x TBE, 130 V | ||
| - | + | ||
[[Image:TUM2010_100521beschriftet.png]] | [[Image:TUM2010_100521beschriftet.png]] | ||
| Line 1,455: | Line 1,029: | ||
{{:Team:TU Munich/Templates/ToggleBoxStart1}}Week08{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/RedBox | text=''in vivo'' constructs }} | {{:Team:TU Munich/Templates/ToggleBoxStart1}}Week08{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/RedBox | text=''in vivo'' constructs }} | ||
{{:Team:TU Munich/Templates/ToggleBoxStart3}} | {{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
| + | |||
===25.05.2010=== | ===25.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]]''' |
** 4 colonies picked from each Plate | ** 4 colonies picked from each Plate | ||
***pSB1K3_HisSig_B0014 | ***pSB1K3_HisSig_B0014 | ||
| Line 1,469: | Line 1,044: | ||
** stained in SybrSafe | ** stained in SybrSafe | ||
<br> | <br> | ||
| - | [[Image:TUM2010_100525beschriftet. | + | [[Image:TUM2010_100525beschriftet.|600px]]<br> |
[[Image:TUM2010_100525bbeschriftet.png]]<br> | [[Image:TUM2010_100525bbeschriftet.png]]<br> | ||
*overnight cultures made of | *overnight cultures made of | ||
| Line 1,477: | Line 1,052: | ||
===26.05.2010=== | ===26.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology #ZYMO RESEARCH DNA Clean&Concentration Kit|Miniprep]]''' of cultures set up [[25.05.2010]] |
**HisSig 3, DL1, DL4 | **HisSig 3, DL1, DL4 | ||
**TrpSig DL2, DL4 | **TrpSig DL2, DL4 | ||
**HisTerm/TrpTerm 1,2,3 | **HisTerm/TrpTerm 1,2,3 | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Restriction|Restriction]]''' |
**analytical: E/P HisSig(3, DL2); TrpSig(DL2, DL4): 1.5 h 37 °C | **analytical: E/P HisSig(3, DL2); TrpSig(DL2, DL4): 1.5 h 37 °C | ||
**prep: E/X HisSig(3, DL2); TrpSig(DL2, DL4): 1.5 h 37 °C | **prep: E/X HisSig(3, DL2); TrpSig(DL2, DL4): 1.5 h 37 °C | ||
**prep: E/S R0011: 1.5 h 37 °C, inactivation 20 min @ 80 °C | **prep: E/S R0011: 1.5 h 37 °C, inactivation 20 min @ 80 °C | ||
| - | ***total volume each 20 | + | ***total volume each 20 µl, 10 µL template |
*'''Gel''': 1% Agarose, TAE - 1,5 h 110 V | *'''Gel''': 1% Agarose, TAE - 1,5 h 110 V | ||
| - | **[[ | + | **[[Team:TU_Munich/Lab#Molecular_Biology #ZYMO RESEARCH Gel DNA Recovery Kit|bands excised]]: all E/X cleaved vectors |
[[Image:TUM2010_100526beschriftet.png]] | [[Image:TUM2010_100526beschriftet.png]] | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Ligation|Ligation]]''': |
**HisSig_3 (E/X) with R0011 (E/S) | **HisSig_3 (E/X) with R0011 (E/S) | ||
**HisSig_DL2 (E/X) with R0011 (E/S) | **HisSig_DL2 (E/X) with R0011 (E/S) | ||
| Line 1,497: | Line 1,072: | ||
**TrpSig_DL4 (E/X) with R0011 (E/S) | **TrpSig_DL4 (E/X) with R0011 (E/S) | ||
***'''batches''' | ***'''batches''' | ||
| - | ***total volume 20 | + | ***total volume 20 µL |
| - | ***2 | + | ***2 µL R0011 (E/S) |
| - | ***2 | + | ***2 µL T4 buffer |
| - | ***1 | + | ***1 µL T4 Ligase |
| - | ***15 | + | ***15 µL vector |
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Transformation|Transformation]]''' of ligations |
** DH5a | ** DH5a | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|PCR]]''' |
**His and TrpSig | **His and TrpSig | ||
**His and TrpTerm | **His and TrpTerm | ||
**R0011 | **R0011 | ||
**B0014 | **B0014 | ||
| - | ***50 | + | ***50 µL total volume<br> |
| - | ***1 | + | ***1 µL template<br> |
| - | ***1 | + | ***1 µl G1004<br> |
| - | ***1 | + | ***1 µl G1005<br> |
| - | ***0.2 | + | ***0.2 µL Taq<br> |
| - | ***5 | + | ***5 µl Taq standard buffer<br> |
***rest water | ***rest water | ||
| + | |||
===27.05.2010=== | ===27.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]]''' |
** 3 colonies picked from each Plate | ** 3 colonies picked from each Plate | ||
***pSB1K3_R0011_HisSig_B0014 (N° 3 from yesterday) | ***pSB1K3_R0011_HisSig_B0014 (N° 3 from yesterday) | ||
| Line 1,597: | Line 1,173: | ||
[[Image:TUM2010_100527bbeschriftet.png]]<br> | [[Image:TUM2010_100527bbeschriftet.png]]<br> | ||
===28.05.2010=== | ===28.05.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology ZYMO_RESEARCH_DNA_Clean.26Concentration_Kit|Miniprep]]''' of cultures from [[27.05.2010]], Elution in 50 uL nuclease free water |
** (1)HisSig 3_1 | ** (1)HisSig 3_1 | ||
** (2)HisSig DL1_3 | ** (2)HisSig DL1_3 | ||
| Line 1,648: | Line 1,224: | ||
<br> | <br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Restriction|analytical digest]]''' (E/P)<br> |
**of all samples. total volume 20 uL, 5 uL template used for Term-constructs, 10 uL teplate for all others | **of all samples. total volume 20 uL, 5 uL template used for Term-constructs, 10 uL teplate for all others | ||
| Line 1,675: | Line 1,251: | ||
:[[Image:TUM2010_100531.png]] | :[[Image:TUM2010_100531.png]] | ||
| - | :Band at ~2100b cut und purified using the [[ | + | :Band at ~2100b cut und purified using the [[Team:TU_Munich/Lab#Molecular_Biology ZYMO RESEARCH Gel DNA Recovery Kit |zymo kit]] |
| - | * [[ | + | * [[Team:TU_Munich/Lab#Molecular_Biology Ligation|ligation]] |
| - | ** HisSig (4 | + | ** HisSig (4 µL of digest) with purified plasmid (with BBa_I719005) |
| - | ** TrpSig (2 | + | ** TrpSig (2 µL of digest) with purified plasmid -=- |
::reason: concentration of His Sig before digest was 1/2 of TrpSig | ::reason: concentration of His Sig before digest was 1/2 of TrpSig | ||
| - | * [[ | + | * [[Team:TU_Munich/Lab#Molecular_Biology Transformation|transformation]] |
: of DH5a with Ligation batches, HisSig1-3, HisSig3-1, TrpSig DL4-1, TrpSig 4-3 | : of DH5a with Ligation batches, HisSig1-3, HisSig3-1, TrpSig DL4-1, TrpSig 4-3 | ||
===01.06.2010=== | ===01.06.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]]''' |
:of Ligations transformed into DH5a yesterday | :of Ligations transformed into DH5a yesterday | ||
| Line 1,793: | Line 1,369: | ||
<br> | <br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Restriction|Restrictions]] ''' |
**psB1A10_TrpTerm/HisTerm with Nsi1, Aat2 | **psB1A10_TrpTerm/HisTerm with Nsi1, Aat2 | ||
**pSB1K3_R0011_HisSig/TrpSig_B0014 with Pst1, Aat2 | **pSB1K3_R0011_HisSig/TrpSig_B0014 with Pst1, Aat2 | ||
| Line 1,802: | Line 1,378: | ||
:psB1A10_TrpTerm/HisTerm and T7 bb dephosphorylated the last 30 min <br> | :psB1A10_TrpTerm/HisTerm and T7 bb dephosphorylated the last 30 min <br> | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Ligation|Ligation]] ''' <br> |
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Transformation|Transformation]] ''' |
<br> liquid culture (10 ml) of pSB1K3_R0011_HisSig/TrpSig_B0014 | <br> liquid culture (10 ml) of pSB1K3_R0011_HisSig/TrpSig_B0014 | ||
| Line 1,812: | Line 1,388: | ||
: checked with blast2seq | : checked with blast2seq | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology PCR|Colony PCR]]''' |
: 8 colonies from each plate of T7His, T7Trp, MonsterHis, MonsterTrp as ligations resulted in many colonies | : 8 colonies from each plate of T7His, T7Trp, MonsterHis, MonsterTrp as ligations resulted in many colonies | ||
| Line 1,830: | Line 1,406: | ||
[[Image:TUM2010_100609_Monster_inverse.jpg|600px]] | [[Image:TUM2010_100609_Monster_inverse.jpg|600px]] | ||
<br> | <br> | ||
| - | gel didn´t work at all --> even after > | + | gel didn´t work at all --> even after > 2 h, bands were not separated correctly, even the 1kb ladder was "stacked" in the gel-pockets, the 100 bp ladder should show EQUAL distances between the lines [http://www.neb.com/nebecomm/productfiles/778/images/N3231_fig1_v1_000034.gif see here], it looks like the gel was "more dense" at the pockets---> no idea what happened --> repeat Monster-digestion tomorrow? |
<br> | <br> | ||
* gel of T7_Plasmid digestion | * gel of T7_Plasmid digestion | ||
| Line 1,845: | Line 1,421: | ||
:Spe1, Aat2 from NEB, 500 U each | :Spe1, Aat2 from NEB, 500 U each | ||
<br> | <br> | ||
| - | * '''[[ | + | * '''[[Team:TU_Munich/Lab#Molecular_Biology Restriction|analytical Digestion]] |
:of Ligation colonies from MonsterHis/trp 1-3 and T7His/Trp 1,2,5/1,2,3 | :of Ligation colonies from MonsterHis/trp 1-3 and T7His/Trp 1,2,5/1,2,3 | ||
:2h digestion | :2h digestion | ||
| - | :: Monster: 6 | + | :: Monster: 6 µL DNA template with Aat2/Spe1 in Buffer 4/Bsa |
| - | :: T7-Signal: 6 | + | :: T7-Signal: 6 µL DNA template with E/P in Buffer 3 |
<br> | <br> | ||
* '''Agarose Gels''' | * '''Agarose Gels''' | ||
| - | :used standards: lmw, 2-log [[ | + | :used standards: lmw, 2-log [[Team:TU_Munich/Lab#Molecular_Biology standards|click here]] |
: Gel1: 1% Agarose in 1xTBE for Digestions of Monsterplasmid | : Gel1: 1% Agarose in 1xTBE for Digestions of Monsterplasmid | ||
:: run in big chamber @ 200 V for 1 h 20 min | :: run in big chamber @ 200 V for 1 h 20 min | ||
| Line 1,875: | Line 1,451: | ||
===14.06.2010=== | ===14.06.2010=== | ||
| - | *'''[[ | + | *'''[[Team:TU_Munich/Lab#Molecular_Biology Ligation|Ligation]]''' |
| - | : 10 | + | : 10 µL pSB1A10_TrpTerm Aat2/Nsi1 0.5 ng/uL |
| - | : 1 | + | : 1 µL R0011_TrpSig_B0014 Aat2/Psb1 11 ng/uL |
| - | : 2 | + | : 2 µL T4 ligase buffer |
| - | : 1 | + | : 1 µL T4 Ligase |
| - | : 6 | + | : 6 µL H2O |
::10 min RT | ::10 min RT | ||
<br> | <br> | ||
| Line 1,896: | Line 1,472: | ||
===15.06.2010=== | ===15.06.2010=== | ||
*Over night cultures | *Over night cultures | ||
| - | *Aliquots of the Promega in vitro expressions kit from | + | *Aliquots of the Promega in vitro expressions kit from ''E. coli'' S30 extract: |
| - | : 40 | + | : 40 µL with aa mix including all aa. |
===16.06.2010=== | ===16.06.2010=== | ||
| Line 1,952: | Line 1,528: | ||
*Miniprep | *Miniprep | ||
| - | : pSBN1A10_TrpSig - 40 | + | : pSBN1A10_TrpSig - 40 µL, 12.5 ng/µL -->very low amount of DNA... |
: culture is slightly red? -> strange because there cannot be any rfp-insert with constitutive promoter as the construct was built up from pSB1A10_TrpTerm (digested) and TrpSignal (PCR product) | : culture is slightly red? -> strange because there cannot be any rfp-insert with constitutive promoter as the construct was built up from pSB1A10_TrpTerm (digested) and TrpSignal (PCR product) | ||
<br> | <br> | ||
*cultures | *cultures | ||
| - | ** | + | **5 ml cultures of DH5a |
::with MonsterTrp, #7,8,10 (Carbamp) | ::with MonsterTrp, #7,8,10 (Carbamp) | ||
:*50 ml cultures of BL21 (DE3) RIL | :*50 ml cultures of BL21 (DE3) RIL | ||
| Line 1,970: | Line 1,546: | ||
: MonsterTrp #7,8,10 | : MonsterTrp #7,8,10 | ||
: positive Control | : positive Control | ||
| - | :: concentrations are too low for sequencing --> again we have to set | + | :: concentrations are too low for sequencing --> again we have to set up 5 ml cultures for tomorrow |
<br> | <br> | ||
* measurements | * measurements | ||
| Line 1,983: | Line 1,559: | ||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| - | {{:Team:TU Munich/Templates/ToggleBoxStart1}}Week13{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/RedBox | text= | + | {{:Team:TU Munich/Templates/ToggleBoxStart1}}Week13{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/RedBox | text=''in vivo'' constructs }} {{:Team:TU_Munich/Templates/BlueBox | text=Testing pSB1A10 }} |
{{:Team:TU Munich/Templates/ToggleBoxStart3}} | {{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
===28.06.2010=== | ===28.06.2010=== | ||
* Sequencing results | * Sequencing results | ||
| - | :psB1A10_TrpSig ( | + | :psB1A10_TrpSig (Positive control) worked: Trp-Sig is inside, directly in front of RFP (without the promotor of the measurement plasmid insert [http://partsregistry.org/Part:BBa_J04450 BBa_J04450], so it seems like everything worked. Furthermore, i performed a promotor prediction with the following tool [http://linux1.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb bacteria promotor prediction tool] to proof if our Trp-Sig in combination with the flanking regions is not forming a promotor,by mischance. According to this, there are two promotors, BUT: <br> one in and after the suffix (so it should be in each of our constructs), but the tools says theres is no known sigma-factor for this promotor! the second one is within the RFP and there is a sigma-factor for this one ( rpoD16). So i don´t see a explanation, why our colonies were pink in contrast to the other "Messplasmids". <br><br> None of the Monsterplasmids contains the signal construct. Probably, the problem is there is no selection methods which allowed us to distinguish uncut plasmids.... --> we should discuss at our next meeting, one possiblity would be connecting our construct to a resistance marker. I summed up all sequences in this document: [[File:25.06.-sequenzierung.doc]] <br><br> |
*Induction in cuvette and measuring fluorescence at the same time IS NOT WORKING! (probably cells are not growing and expressing very well, maybe lacking oxygen despite stirring. Bleaching is more unlikely) <br> In vivo, measuring plasmid (at least GFP) works! we optimized the paramters for fluorescence measurment! We tried different OD´s and found out that only measurments below OD 0.4 result in meaningful measurements. <br> as a result, in vitro expression did somehow not work, reasons are unclear, maybe too low DNA-concentrations. <br> positvie control psB1A10_TrpSig was pink again, we have to wait the results from GATC | *Induction in cuvette and measuring fluorescence at the same time IS NOT WORKING! (probably cells are not growing and expressing very well, maybe lacking oxygen despite stirring. Bleaching is more unlikely) <br> In vivo, measuring plasmid (at least GFP) works! we optimized the paramters for fluorescence measurment! We tried different OD´s and found out that only measurments below OD 0.4 result in meaningful measurements. <br> as a result, in vitro expression did somehow not work, reasons are unclear, maybe too low DNA-concentrations. <br> positvie control psB1A10_TrpSig was pink again, we have to wait the results from GATC | ||
| Line 2,001: | Line 1,577: | ||
:using Invitrogen kit | :using Invitrogen kit | ||
:measuring kinetics for 3 h @ 37°C | :measuring kinetics for 3 h @ 37°C | ||
| - | :40 | + | :40 µL + 5 µL pSB1A10_TrpSig (126 ng/µL) + 0.5 µL 100x L-Arabinose (=0.2%) + 4.5 µL H2O |
| - | ::observations: | + | ::observations: GFP signal grows, after 30 min it crashes. RFP grows |
| - | ::emission spectra for | + | ::emission spectra for GFP and RFP result in no spectrum |
::looks strange, a problem might be evaporation of liquid and hence scattering of light which produces artefacts | ::looks strange, a problem might be evaporation of liquid and hence scattering of light which produces artefacts | ||
<br> | <br> | ||
| Line 2,012: | Line 1,588: | ||
===02.07.2010=== | ===02.07.2010=== | ||
*Miniprep | *Miniprep | ||
| - | :pSB1A10_XS: 30 | + | :pSB1A10_XS: 30 µL 10 ng/µL |
| - | :pSB1A10_TrpSig: 30 | + | :pSB1A10_TrpSig: 30 µL 10 ng/µL |
<br> | <br> | ||
*Transformation | *Transformation | ||
| Line 2,019: | Line 1,595: | ||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| - | {{:Team:TU Munich/Templates/ToggleBoxStart1}}Week15{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/ClearBox }} {{:Team:TU_Munich/Templates/BlueBox | text=Testing | + | {{:Team:TU Munich/Templates/ToggleBoxStart1}}Week15{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/ClearBox }} {{:Team:TU_Munich/Templates/BlueBox | text=Testing pSB1A10 }} {{:Team:TU_Munich/Templates/ClearBox }} |
{{:Team:TU Munich/Templates/ToggleBoxStart3}} | {{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
===05.07.2010=== | ===05.07.2010=== | ||
| Line 2,031: | Line 1,607: | ||
===06.07.2010=== | ===06.07.2010=== | ||
*'''In vivo measurement''' | *'''In vivo measurement''' | ||
| - | : BL21 | + | : BL21 pSB1A10_XS - positive control (to check the measurement plasmid...) |
: GFP, RFP Fluorescence | : GFP, RFP Fluorescence | ||
:induced with 0.2% arabinose in (1), uninduced (2) | :induced with 0.2% arabinose in (1), uninduced (2) | ||
| Line 2,077: | Line 1,653: | ||
**Description: mCherry cut was ok, Plasmid was cut at least once (linear DNA), generally contaminated with genomic DNA | **Description: mCherry cut was ok, Plasmid was cut at least once (linear DNA), generally contaminated with genomic DNA | ||
*Ligation | *Ligation | ||
| - | ** | + | **50 ng Plasmid and 34 ng Insert |
**ca. 30min @ RT | **ca. 30min @ RT | ||
*Transformation of DH5a cells with ligation samples | *Transformation of DH5a cells with ligation samples | ||
| Line 2,093: | Line 1,669: | ||
*analytic gel of Mini preps and ligation of the previous day | *analytic gel of Mini preps and ligation of the previous day | ||
**preps still hold genomic DNA | **preps still hold genomic DNA | ||
| - | **mCherry Plasmid runs at ca. | + | **mCherry Plasmid runs at ca. 2400 bp |
*Digestion | *Digestion | ||
| Line 2,103: | Line 1,679: | ||
*Ligation | *Ligation | ||
**Plasmid (from previous day) and mCherry-Insert | **Plasmid (from previous day) and mCherry-Insert | ||
| - | **ca. | + | **ca. 30 min @ RT |
*Transformation of DH5a cells with | *Transformation of DH5a cells with | ||
**ligation samples (=> no colonies the next day) | **ligation samples (=> no colonies the next day) | ||
| Line 2,117: | Line 1,693: | ||
*analytic gel of Mini preps and ligation of the previous day | *analytic gel of Mini preps and ligation of the previous day | ||
**preps still hold genomic DNA | **preps still hold genomic DNA | ||
| - | **mCherry Plasmid runs at ca. | + | **mCherry Plasmid runs at ca. 2400 bp |
**Gel (1%) | **Gel (1%) | ||
[[Image:TUM2010_100812 ligation100811 mChPrep.jpg]] | [[Image:TUM2010_100812 ligation100811 mChPrep.jpg]] | ||
| Line 2,132: | Line 1,708: | ||
*Ligation | *Ligation | ||
**Plasmid and mCherry-Insert | **Plasmid and mCherry-Insert | ||
| - | **ca. | + | **ca. 30 min @ RT |
*Transformation of DH5a cells with | *Transformation of DH5a cells with | ||
**ligation samples (=> no colonies the next day) | **ligation samples (=> no colonies the next day) | ||
| Line 2,181: | Line 1,757: | ||
*Ligation | *Ligation | ||
**Plasmid (from earlier date) and mCherry-Insert ('''from 11.08''') | **Plasmid (from earlier date) and mCherry-Insert ('''from 11.08''') | ||
| - | **ca. | + | **ca. 30 min @ RT |
**'''used new ligase and new ligase buffer''' | **'''used new ligase and new ligase buffer''' | ||
| Line 2,221: | Line 1,797: | ||
*Ligation | *Ligation | ||
**Plasmid (from previous day) and mCherry-Insert | **Plasmid (from previous day) and mCherry-Insert | ||
| - | **ca. | + | **ca. 30 min @ RT |
*Transformation of DH5a cells with | *Transformation of DH5a cells with | ||
**ligation samples (=> no colonies the next day) | **ligation samples (=> no colonies the next day) | ||
| Line 2,260: | Line 1,836: | ||
*Ligation | *Ligation | ||
**Plasmid His-Term(Trp-Term) and Linker | **Plasmid His-Term(Trp-Term) and Linker | ||
| - | **ca. | + | **ca. 30 min @ RT |
*Transformation of DH5a cells with | *Transformation of DH5a cells with | ||
**ligation samples | **ligation samples | ||
| Line 2,497: | Line 2,073: | ||
===31.08.2010=== | ===31.08.2010=== | ||
*'''Fluorescence measurement (positive control experiment):''' | *'''Fluorescence measurement (positive control experiment):''' | ||
| - | **Settings: GFP-Excitation: | + | **Settings: GFP-Excitation: 501 nm; mCherry-Excitation: 587 nm; |
**endpoint measurements of: | **endpoint measurements of: | ||
| - | ***Timepoints of measurement: | + | ***Timepoints of measurement: 3 h after induction and 9 h after induction (1.5 h and 7 h for 15x-sample) |
***Samples: | ***Samples: | ||
****pSB1A10mod-mCherry (27b) in BL21 cells, induced with ca. 0.4% L-Arabinose | ****pSB1A10mod-mCherry (27b) in BL21 cells, induced with ca. 0.4% L-Arabinose | ||
| Line 2,541: | Line 2,117: | ||
*'''Fluorescence measurement (positive control experiment):''' | *'''Fluorescence measurement (positive control experiment):''' | ||
**endpoint measurements: | **endpoint measurements: | ||
| - | ***Timepoints of measurement: | + | ***Timepoints of measurement: 3 h after induction and 9 h after induction (1.5 h and 7 h for 15x-smaple) |
| - | ***Settings: GFP-Excitation: | + | ***Settings: GFP-Excitation: 501 nm; mCherry-Excitation: 587 nm; RFP-Excitation: 584 nm |
***Samples: | ***Samples: | ||
****pSB1A10mod-mCherry (32a) in DH5a cells, induced with ca. 0.4% L-Arabinose | ****pSB1A10mod-mCherry (32a) in DH5a cells, induced with ca. 0.4% L-Arabinose | ||
| Line 2,561: | Line 2,137: | ||
*'''Amplifing''' the Arabinose-inducable promotor pBAD | *'''Amplifing''' the Arabinose-inducable promotor pBAD | ||
| - | **Resuspending the BioBrick I13453 with | + | **Resuspending the BioBrick I13453 with 10 µl in well 1F in the 2010 Distribution |
| - | **PCR using | + | **PCR using 1 µl template (programm "igempcr") |
**Purified using DNA Concentrator (ZymoKit) | **Purified using DNA Concentrator (ZymoKit) | ||
| - | *Digestion (EcoRI and SpeI) of PCR product and heat inactivation ( | + | *Digestion (EcoRI and SpeI) of PCR product and heat inactivation (20 min @ 80°C) |
*Digestion of the target vectors using EcoRI and XbaI | *Digestion of the target vectors using EcoRI and XbaI | ||
| Line 2,580: | Line 2,156: | ||
:=> '''Interpretation:''' Gel is overloaded! However digestion seemed to work since the bands show correct masses. | :=> '''Interpretation:''' Gel is overloaded! However digestion seemed to work since the bands show correct masses. | ||
| - | *Extraction of bands at ca. | + | *Extraction of bands at ca. 6000 bp |
| - | *10 µl '''ligation''' of | + | *10 µl '''ligation''' of 50 ng of each digested vector with 8ng insert |
| - | *'''Transformation''' of DH5a cells using | + | *'''Transformation''' of DH5a cells using 8 µl ligation sample |
===03.09.2010=== | ===03.09.2010=== | ||
*Colony PCR | *Colony PCR | ||
| Line 2,624: | Line 2,200: | ||
**Induction with 0.2% L-arabinose | **Induction with 0.2% L-arabinose | ||
**Measurement of OD600 | **Measurement of OD600 | ||
| - | **Fluorescence measurement at | + | **Fluorescence measurement at 30 min, 150 min (only Pos.Control) and 4.5 h |
**Samples: | **Samples: | ||
::pSB1A10mod_Pbad_mCherry | ::pSB1A10mod_Pbad_mCherry | ||
| Line 2,648: | Line 2,224: | ||
**Induction with 0.2% L-arabinose | **Induction with 0.2% L-arabinose | ||
**Measurement of OD600 | **Measurement of OD600 | ||
| - | **Fluorescence measurement at | + | **Fluorescence measurement at 24 h and different OD600 |
**Samples: | **Samples: | ||
::pSB1A10mod_Pbad_mCherry | ::pSB1A10mod_Pbad_mCherry | ||
| - | -->OD600 0.05 reasonable for our cell measurements. Positive control works fine after | + | -->OD600 0.05 reasonable for our cell measurements. Positive control works fine after 24 h induction. |
[[Image:TUM2010_Graph2.jpg|600px]] | [[Image:TUM2010_Graph2.jpg|600px]] | ||
| Line 2,674: | Line 2,250: | ||
***pSB1A10mod_pBAD_TrpTerm_mCherry_TrpSignal | ***pSB1A10mod_pBAD_TrpTerm_mCherry_TrpSignal | ||
**Induction with 0.4% L-arabinose and 1mM IPTG | **Induction with 0.4% L-arabinose and 1mM IPTG | ||
| - | **Timepoints: | + | **Timepoints: 1 h, 2 h, 4 h, 10 h, 16 h (induced the day before) |
::(OD checked seperately each time; average ODs=0.02-0.06) | ::(OD checked seperately each time; average ODs=0.02-0.06) | ||
| Line 2,685: | Line 2,261: | ||
===10.09.2010=== | ===10.09.2010=== | ||
*Fluorescence measurements | *Fluorescence measurements | ||
| - | **Induction with 0.4% L-arabinose and | + | **Induction with 0.4% L-arabinose and 1 mM IPTG |
**Measurement of OD600 | **Measurement of OD600 | ||
| - | **Fluorescence measurement at | + | **Fluorescence measurement at 15 h and 26 h |
*Samples: | *Samples: | ||
::pSB1A10mod_Pbad_mCherry - Positive Control | ::pSB1A10mod_Pbad_mCherry - Positive Control | ||
| Line 2,699: | Line 2,275: | ||
*Conclusion: | *Conclusion: | ||
| - | His-Switch DOES NOT seem to work!!! Measurements of Trp-Switch look weird, since Ara-induced culture showed strong mCherry signal than Ara+IPTG-induced culture. Maybe growing condition were not sufficient. We will use | + | His-Switch DOES NOT seem to work!!! Measurements of Trp-Switch look weird, since Ara-induced culture showed strong mCherry signal than Ara+IPTG-induced culture. Maybe growing condition were not sufficient. We will use 50 ml flask next time! |
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| Line 2,710: | Line 2,286: | ||
*<i>In vitro</i> Kit measurement: | *<i>In vitro</i> Kit measurement: | ||
**Test of <i>In vitro</i> kit with positive control | **Test of <i>In vitro</i> kit with positive control | ||
| - | ** | + | **1 h on 37°C |
::--> no significant mCherry signal detectable. <i>In vitro</i> Kit measurements aborted. | ::--> no significant mCherry signal detectable. <i>In vitro</i> Kit measurements aborted. | ||
| Line 2,722: | Line 2,298: | ||
:*Growth on 37°C until OD600 = 0.6, then induction and growth on 25°C over night | :*Growth on 37°C until OD600 = 0.6, then induction and growth on 25°C over night | ||
| - | :*each sample uininduced, induced with 0.4% L-arabinose and 0.4% L-arabinose + | + | :*each sample uininduced, induced with 0.4% L-arabinose and 0.4% L-arabinose + 1 mM IPTG,respectively |
* 30% Glycerol stock in LB | * 30% Glycerol stock in LB | ||
| Line 2,731: | Line 2,307: | ||
**Induction with 0.4% L-arabinose and 1mM IPTG | **Induction with 0.4% L-arabinose and 1mM IPTG | ||
**Measurement of OD600 | **Measurement of OD600 | ||
| - | **Fluorescence measurement at | + | **Fluorescence measurement at 16 h |
*Samples: | *Samples: | ||
::pSB1A10mod_Pbad_mCherry - Positive Control | ::pSB1A10mod_Pbad_mCherry - Positive Control | ||
| Line 2,805: | Line 2,381: | ||
**Trp-Switch EcoRI/PstI | **Trp-Switch EcoRI/PstI | ||
**His-Switch EcoRI/PstI | **His-Switch EcoRI/PstI | ||
| - | ::--> Heat inactivation | + | ::--> Heat inactivation 20 min, 80°C |
*Ligation | *Ligation | ||
| Line 2,859: | Line 2,435: | ||
| - | * | + | *10 µl ligations using digested samples from [[21.09.2010]] |
**pSB1A2_R0011 (SpeI/PstI cut) + HisSig (XbaI/PstI cut) | **pSB1A2_R0011 (SpeI/PstI cut) + HisSig (XbaI/PstI cut) | ||
**pMalachiteApt (EcoRI/PstI cut) + TrpSwitch (EcoRI/PstI cut) | **pMalachiteApt (EcoRI/PstI cut) + TrpSwitch (EcoRI/PstI cut) | ||
| Line 2,873: | Line 2,449: | ||
::tac_BB1006Switch_MalachitApt + tac_BB1006Signal (both PCR-amplified without BB0014 Terminator) | ::tac_BB1006Switch_MalachitApt + tac_BB1006Signal (both PCR-amplified without BB0014 Terminator) | ||
:*Sample Mix: | :*Sample Mix: | ||
| - | :: | + | ::2 µl sigma70 satured RNA <i>E.Coli</i> Polymerase (epicenter Biozyme) (2U) |
::ca. 1µg DNA template | ::ca. 1µg DNA template | ||
| - | :: | + | ::5 µM Malachite Green |
| - | :: | + | ::10 µM DTT |
| - | :: | + | ::4 µM NTPs |
| - | :: | + | ::40 mM Tris-HCl pH = 7.1 @ 37°C; 7.5 @ 22°C |
| - | :: | + | ::10 mM MgCl2 |
| - | :: | + | ::150 mM KCl |
| - | ::added H20 to total volume of | + | ::added H20 to total volume of 100 µl |
:*Kinetics measurement @ 37°C | :*Kinetics measurement @ 37°C | ||
| - | ::*Excitation | + | ::*Excitation 630 nm |
| - | ::*Emission | + | ::*Emission 655 nm |
[[Image:TUM2010_100924 MalachitKineticsPosControl BB1006.jpg|600px]] | [[Image:TUM2010_100924 MalachitKineticsPosControl BB1006.jpg|600px]] | ||
::*Results: | ::*Results: | ||
| Line 2,890: | Line 2,466: | ||
:*Scanning measurements | :*Scanning measurements | ||
| - | ::*Exitation: | + | ::*Exitation: 630 nm |
::*Results: | ::*Results: | ||
:::no detactable peaks between 640nm and 800nm in any sample at any measured time (Settings were checked by detecting scattering peaks). | :::no detactable peaks between 640nm and 800nm in any sample at any measured time (Settings were checked by detecting scattering peaks). | ||
| Line 2,929: | Line 2,505: | ||
* PCR of switch_phi_T7-construct to obtain dsDNA (conditions: igemPCR and 30s elongation time). Samples were purified using Qiagen MinElute. | * PCR of switch_phi_T7-construct to obtain dsDNA (conditions: igemPCR and 30s elongation time). Samples were purified using Qiagen MinElute. | ||
| - | * 1th measurment: <br> Maxi´s NEB buffer conditions: | + | * 1th measurment: <br> Maxi´s NEB buffer conditions: 40 mM Tris pH 7.4 @ RT, 40mM Mg2Cl, 5 µM Malachit-green, 4 mM NTPs, 2,5 U RNA-Polymerase <br>All DNA templates were all added to a final concentration of 200nM. |
**sample 1: Positive control | **sample 1: Positive control | ||
**sample 2: negative control (= switch without any signal) ('''new DTT''' used for this sample) | **sample 2: negative control (= switch without any signal) ('''new DTT''' used for this sample) | ||
| Line 2,942: | Line 2,518: | ||
* 2th measurment: | * 2th measurment: | ||
| - | **"Paper´s" buffer conditions: | + | **"Paper´s" buffer conditions: 40 mM Tris pH 7.9 @ RT, 6mM Mg2Cl, 5 µM Malachit-green, 100 mM KCl, 0.8 mM NTPs, 2,5 U RNA-Polymerase |
***sample 1: Positive control | ***sample 1: Positive control | ||
***sample 2: negative control (= switch without signal) | ***sample 2: negative control (= switch without signal) | ||
| - | **Maxi´s NEB buffer conditions: | + | **Maxi´s NEB buffer conditions: 40 mM Tris, 40 mM Mg2Cl, 10 µM Malachit-green, ph 7.4 @ RT |
***sample 3: Positive control ('''new DTT''' used for this sample) | ***sample 3: Positive control ('''new DTT''' used for this sample) | ||
***sample 4: Negative control (= switch without signal) | ***sample 4: Negative control (= switch without signal) | ||
| Line 2,970: | Line 2,546: | ||
::2.5 U T7 RNA Polymerase | ::2.5 U T7 RNA Polymerase | ||
:*Results: | :*Results: | ||
| + | :[[Image:TUM2010_10104_Kinetik.jpg|600px]] | ||
:Fluorescence increased in all samples in a very similar way, suggesting all signal to work equally well. | :Fluorescence increased in all samples in a very similar way, suggesting all signal to work equally well. | ||
| Line 2,994: | Line 2,571: | ||
::2.5 U T7 RNA Polymerase | ::2.5 U T7 RNA Polymerase | ||
:*Results: | :*Results: | ||
| + | :[[Image:TUM2010_10104_Kinetik2.jpg|600px]] | ||
::*Very strange results!! Both negative controls increased strongly compared to the positive controls that only slightly increased (buffer A seems to be better for transcription than buffer B). | ::*Very strange results!! Both negative controls increased strongly compared to the positive controls that only slightly increased (buffer A seems to be better for transcription than buffer B). | ||
:::=> Maybe something went very wrong. However, last Friday we observed a similar, strange behavior. Maybe the positive control is no good choice. It would make sence to use T7-Switch+SignalA_1a as a reference. | :::=> Maybe something went very wrong. However, last Friday we observed a similar, strange behavior. Maybe the positive control is no good choice. It would make sence to use T7-Switch+SignalA_1a as a reference. | ||
::*Signals were generally stronger. 10 µM of malachite green seems to be quite good. | ::*Signals were generally stronger. 10 µM of malachite green seems to be quite good. | ||
| + | |||
| + | ===05.10.2010=== | ||
| + | * T7 ''in vitro measurements'' | ||
| + | |||
| + | ** First measurement of the day: Signal, 1d, 2c, 3b | ||
| + | *** Master Mix: | ||
| + | *** 204 µl 2x Buffer | ||
| + | *** 40.8 µl DTT | ||
| + | *** 8.16 µl NTPs | ||
| + | *** 21.05 µl Switch (1.94 µM) | ||
| + | *** signals: # volume µl | ||
| + | *** negative control - 1.213 | ||
| + | *** 1d - 1.290 | ||
| + | *** 2c - 1.450 | ||
| + | *** 3b - 1.376 | ||
| + | [[Image:TUM2010_10105_Kinetik1.jpg|600px]] | ||
| + | |||
| + | ** PCR with positive control and T7 switch | ||
| + | *** Mastermix: | ||
| + | PCR T7 switch/positive signal | ||
| + | ** 32 x Mastermix: 1600 µl total volume | ||
| + | *** 32 µl dNTPs | ||
| + | *** 32 µl T7 forward primer | ||
| + | *** 32 µl T7 reverser primer | ||
| + | *** 160 µl 10x buffer | ||
| + | *** 96 µl MgCl2 | ||
| + | *** 6.4 µl Taq Polymerase | ||
| + | *** 10 µl Template (some older PCR) | ||
| + | *** 1209.6 µl H2O | ||
| + | |||
| + | ** Second measurement of the day: Signal negative control, nonsense signal, 1a (with 400 µM signal), 1a (with 2 µM signal) | ||
| + | *** *** Master Mix: | ||
| + | *** 204 µl 2x Buffer | ||
| + | *** 40.8 µl DTT | ||
| + | *** 8.16 µl NTPs | ||
| + | *** 21.05 µl Switch (1.94 µM) | ||
| + | *** signals: # volume µl | ||
| + | *** negative control - 30.34 | ||
| + | *** 1d - 1.213 | ||
| + | *** 2c - 2.672 | ||
| + | *** 3b - 13.36 | ||
| + | |||
| + | [[Image:TUM2010_10105_Kinetik2.jpg|600px]] | ||
| + | |||
| + | ** Evaluation of different malachite green concentrations | ||
| + | *** 5 µM, 10 µM, 15 µM, 25 µM chosen | ||
| + | *** high temperature dependency | ||
| + | *** Equilibration of samples important | ||
| + | |||
| + | ===06.10.2010=== | ||
| + | * PCR | ||
| + | ** yesterday's PCR Purification using Zymo Clear and Concentrated | ||
| + | *** yields | ||
| + | *** Switch: 164 ng/µl | ||
| + | *** positive control: 28 ng/µl | ||
| + | *** problems with the temperature? Yield too low! | ||
| + | |||
| + | ** PCR of positive control | ||
| + | *** annealing temperature set to 48°C | ||
| + | *** 20 x Mastermix: | ||
| + | *** 20 µl dNTPs | ||
| + | *** 20 µl T7 forward primer | ||
| + | *** 20 µl T7 reverser primer | ||
| + | *** 100 µl 10x buffer | ||
| + | *** 60 µl MgCl2 | ||
| + | *** 4 µl Taq Polymerase | ||
| + | *** 1 µl Template (some older PCR) | ||
| + | *** 775 µl H2O | ||
| + | |||
| + | ===07.10.2010=== | ||
| + | * T7 Trancription | ||
| + | ** new buffer: | ||
| + | *** Stocks, volume for 20 ml, endconcentration in 1x | ||
| + | *** 1M Tris/HCl, 1.6 ml, 40 mM | ||
| + | *** 500 mM MgCl2, 3.2 ml, 40 mM | ||
| + | *** 250 mM malachite green, 1.6 ml, 20 µM | ||
| + | *** H20 | ||
| + | ** Measurement | ||
| + | *** 4.1 x, switch, switch + nonsense, switch + 1a, switch + 1c | ||
| + | *** 21.16 µl switch, 1.93 µM | ||
| + | *** 10.25 µl RPO | ||
| + | *** 205 µl buffer | ||
| + | *** 41 µl DTT 100 µM | ||
| + | *** 20.5 µl rNTPs | ||
| + | *** H2O | ||
| + | |||
| + | [[Image:TUM2010_10107_Kinetik1.jpg|600px]] | ||
| + | |||
| + | **second measurement | ||
| + | ** Measurement | ||
| + | *** 4.1 x, switch, switch + nonsense, positive control + 1b, positive control + 1b | ||
| + | *** 21.16 µl switch, 1.93 µM | ||
| + | *** 10.25 µl RPO | ||
| + | *** 205 µl buffer | ||
| + | *** 41 µl DTT 100 µM | ||
| + | *** 20.5 µl rNTPs | ||
| + | *** H2O | ||
| + | |||
| + | |||
| + | [[Image:TUM2010_10107_Kinetik2.jpg|600px]] | ||
| + | |||
| + | ===08.10.2010=== | ||
| + | |||
| + | |||
| + | [[Image:TUM2010_101008_Kinetik2.jpg|600px]] | ||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| Line 3,004: | Line 2,687: | ||
{{:Team:TU Munich/Templates/ToggleBoxStart1}}Week29{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/ClearBox | text=Cloning }} {{:Team:TU_Munich/Templates/ClearBox | text=Measurements }} {{:Team:TU_Munich/Templates/GreenBox | text=T7 Switch }} | {{:Team:TU Munich/Templates/ToggleBoxStart1}}Week29{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/ClearBox | text=Cloning }} {{:Team:TU_Munich/Templates/ClearBox | text=Measurements }} {{:Team:TU_Munich/Templates/GreenBox | text=T7 Switch }} | ||
{{:Team:TU Munich/Templates/ToggleBoxStart3}} | {{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
| + | |||
===11.10.2010=== | ===11.10.2010=== | ||
* PCR T7 switch | * PCR T7 switch | ||
| Line 3,038: | Line 2,722: | ||
** Voltage set to 1000 (maximum) | ** Voltage set to 1000 (maximum) | ||
** 38°C, every 15 s measurement | ** 38°C, every 15 s measurement | ||
| + | **[[Image:TUM2010_101011_Kinetik.jpg|600px]] | ||
** start at 5 au, end at 25 au | ** start at 5 au, end at 25 au | ||
** rise visible over time in all samples, nonsense signal with the fastest rise, 1a similiar, end signal similiar in all measurements | ** rise visible over time in all samples, nonsense signal with the fastest rise, 1a similiar, end signal similiar in all measurements | ||
| Line 3,161: | Line 2,846: | ||
===14.10.2010=== | ===14.10.2010=== | ||
* yesterday's measurement | * yesterday's measurement | ||
| + | **[[Image:TUM2010_101014_Kinetik.jpg|600px]] | ||
** rise visible, comparable to tuesday measurement | ** rise visible, comparable to tuesday measurement | ||
** samples frozen, put on 15 % PAGE | ** samples frozen, put on 15 % PAGE | ||
| Line 3,194: | Line 2,880: | ||
:--> yesterdays results bad because no switch :) | :--> yesterdays results bad because no switch :) | ||
| - | *15 % | + | *15 % 6 M urea PAGE |
[[Image:TUM2010 PAGE 101014.png|600px]] | [[Image:TUM2010 PAGE 101014.png|600px]] | ||
| Line 3,665: | Line 3,351: | ||
*** 1.33 µl backbone (I thought 50 ng, actually 25 ng) | *** 1.33 µl backbone (I thought 50 ng, actually 25 ng) | ||
*** 0.23 µl insert, 1:10 diluted | *** 0.23 µl insert, 1:10 diluted | ||
| - | *** 17.37 | + | *** 17.37 µl H2O |
*** incubated till transformation | *** incubated till transformation | ||
| Line 3,823: | Line 3,509: | ||
===23.10.2010=== | ===23.10.2010=== | ||
| + | *''In vitro'' transcription using ''E. coli'' RPO | ||
| + | ** PCR of HisSig, TrpSig, HisTerm, TrpTerm, 16z (positive control) | ||
| + | *** 100 µl per template, 800 µl total | ||
| + | *** 16 µl dNTPs | ||
| + | *** 16 µl forward primer | ||
| + | *** 16 µl reverse primer | ||
| + | *** 80 µl Taq-buffer Mg-free | ||
| + | *** 3.2 µl Taq Polymerase | ||
| + | *** 48 µl MgCl2 | ||
| + | *** 5 µl template per 100 µl | ||
| + | *** 615.8 µl H2O | ||
| + | ** PCR Purification using DNA Quick and Clean | ||
| + | *** yield: Template - # - concentration [ng/µl] | ||
| + | *** 16z - 1 - 146 | ||
| + | *** 16z - 2 - 138 | ||
| + | *** 16z - 3 - 138 | ||
| + | *** 16z - 4 - 128 | ||
| + | *** HisTerm - 1 - 96 | ||
| + | *** HisTerm - 2 - 160 | ||
| + | *** TrpTerm - 1 - 204 | ||
| + | *** TrpTerm - 2 - 190 | ||
| + | *** HisSig - 1 - 160 | ||
| + | *** HisSig - 2 - 165 | ||
| + | *** TrpSig - 1 - 83.5 | ||
| + | *** TrpSig - 2 - 166 | ||
| + | **Measurement | ||
| + | *** 16z positive control, TrpTerm, both with and without signal | ||
| + | *** 4.1 x Master Mix | ||
| + | *** 105 µl Buffer | ||
| + | *** 41 µl DTT | ||
| + | *** 10.25 µl RPO | ||
| + | *** 10.25 µl rNTPs | ||
| + | *** 77.9 µl H2O | ||
| + | *** take 84 µl Master Mix | ||
| + | *** add to: | ||
| + | *** 10.85 µl 16 z + 6.1 µl H20 | ||
| + | *** 10.85 µl 16 z + 6.1 µl TrpSig | ||
| + | *** 10.92 µl TrpTerm + 6.1 µl H20 | ||
| + | *** 10.92 µl TrpTerm + 6.1 µl TrpSig | ||
| + | [[Image:TUM2010_101023_kinetik.png|600px]] | ||
| + | Only the positive control without Signal increases! | ||
| + | |||
| + | |||
| + | * Cloning of MPA into pSB1C3 | ||
| + | ** Checked plates, no clones | ||
| + | ** being sad | ||
| + | ** said stupid plates | ||
| + | ** Repetition of ligation | ||
| + | *** for 50 ng plasmid backbone | ||
| + | *** 2.6 µl pBS1C3 E/P digested | ||
| + | *** 0.23 µl MPA, E/P digested | ||
| + | *** 2 µl T4 ligase buffer - I checked four buffers, all were precipitated, took a completely new one and aliquoted it! | ||
| + | *** 1 µl T4 Ligase | ||
| + | *** for 100 ng plasmid backbone | ||
| + | *** 5.2 µl pBS1C3 E/P digested | ||
| + | *** 0.46 µl MPA, E/P digested | ||
| + | *** 2 µl T4 ligase buffer - I checked four buffers, all were precipitated, took a completely new one and aliquoted it! | ||
| + | *** 1 µl T4 Ligase | ||
| + | *** transformation into pSB1K3 (just in case...) | ||
| + | *** 4.5 µl pSB1K3 E/P digested | ||
| + | *** 0.23 µl MPA, E/P digested | ||
| + | *** 2 µl T4 ligase buffer - I checked four buffers, all were precipitated, took a completely new one and aliquoted it! | ||
| + | *** 1 µl T4 Ligase | ||
| + | ** found some pSB1C3_RFP clones in the evening, 5 ml cultures | ||
| + | ** stroke over the plate with a pipette tip, 5 ml culture | ||
===24.10.2010=== | ===24.10.2010=== | ||
| + | *Cloning of MPA into pSB1C3 | ||
| + | ** Miniprep of pSB1C3_RFP and something random from the plate of no clones | ||
| + | *** mixed up samples: yields were good, but I guess they don't matter | ||
| + | ** very tiny clones found in the morning | ||
| + | ** slightly bigger ones by noon | ||
| + | ** pickable clones by evening | ||
| + | ** picked 10 clones, 5 ml cultures | ||
| + | |||
| + | * BBa_K494001-BBa_K494006 | ||
| + | ** 2x 5 ml LB_Amp from glycerin stocks | ||
| + | |||
| + | * Measurements with T7 RPO and ''E. coli'' constructs | ||
| + | ** 16z positive control, TrpTerm, both with and without signal | ||
| + | *** 4.1 x Master Mix | ||
| + | *** 205 µl Buffer | ||
| + | *** 41 µl DTT | ||
| + | *** 10.25 µl RPO | ||
| + | *** 10.25 µl rNTPs | ||
| + | *** 77.9 µl H2O | ||
| + | |||
| + | *** take 76.2 µl Master Mix | ||
| + | *** add to: | ||
| + | *** 10.07 µl 16 z + 13.7 µl H20 | ||
| + | *** 10.07 µl 16 z + 9.61 µl TrpSig + 4 µl H2O | ||
| + | *** 23.67 µl HisTerm (1) | ||
| + | *** 14.2 µl HisTerm (2) + 9.61 µl TrpSig | ||
| + | |||
| + | [[Image:TUM2010_101024_kinetik.JPG|600px]] | ||
| + | It looks like an increase in 3 of 4 traces, but spectr show no sign of malachite green binding: | ||
| + | [[Image:TUM2010_101024_scan.JPG|600px]] | ||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| Line 3,834: | Line 3,615: | ||
{{:Team:TU Munich/Templates/ToggleBoxStart3}} | {{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
The Week of Wiki freeze! | The Week of Wiki freeze! | ||
| + | |||
| + | ===25.10.2010=== | ||
| + | * Submitting Parts | ||
| + | ** Miniprep of yesterday's 5 ml cultures using Zymo Classic | ||
| + | ** MPA into pSB1C3 | ||
| + | *** # - concentration (ng/µl) | ||
| + | *** 1 - 45.5 | ||
| + | *** 2 - 96.5 | ||
| + | *** 3 - 19.5 | ||
| + | *** 4 - 31 | ||
| + | *** 5 - 122 | ||
| + | *** 6 - 164 | ||
| + | *** 7 - 30 | ||
| + | *** 8 - 39.5 | ||
| + | *** 9 - 20.5 | ||
| + | *** 10 - 113 | ||
| + | |||
| + | ** BBa_K494001-BBa_K494006 | ||
| + | *** BioBrick - # - concentration in ng/µl | ||
| + | *** BBa_K494001 - 1 - 179 | ||
| + | *** BBa_K494001 - 2 - 182 | ||
| + | *** BBa_K494002 - 1 - 214 | ||
| + | *** BBa_K494002 - 2 - 288 | ||
| + | *** BBa_K494003 - 1 - 160 | ||
| + | *** BBa_K494003 - 2 - 202 | ||
| + | *** BBa_K494004 - 1 - 308 | ||
| + | *** BBa_K494004 - 2 - 322 | ||
| + | *** BBa_K494005 - 1 - 290 | ||
| + | *** BBa_K494005 - 2 - 126 | ||
| + | *** BBa_K494006 - 1 - 290 | ||
| + | *** BBa_K494006 - 2 - 232 | ||
| + | |||
| + | ** control digestion of everything | ||
| + | *** EcoRI/PstI | ||
| + | *** MPA clones 1, 3, 4, 7, 8, 9: 8 µl | ||
| + | *** all other: 4 µl | ||
| + | *** 7 x for 4 µl | ||
| + | *** 14 µl 10 x BSA | ||
| + | *** 14 µl 10 NEB3 | ||
| + | *** 2.1 µl EcoRI HF | ||
| + | *** 2.1 µl PstI | ||
| + | *** water as needed | ||
| + | |||
| + | *** 19 x for 4 µl | ||
| + | *** 38 µl 10x BSA | ||
| + | *** 38 µl 10x NEB3 | ||
| + | *** 5.7 µl EcoRI HF | ||
| + | *** 5.7 µl PstI | ||
| + | *** water as needed | ||
| + | |||
| + | *** 37°C, 1 hour, fitted just perfectly into the heat block | ||
| + | |||
| + | *** recognized later that MPA 2, 5, 6, 10 were digested without DNA | ||
| + | *** NotI Digestion with --> PstI empty... Time to finish :) | ||
| + | *** 10 µl NEB | ||
| + | *** 10 µl 10x BSA | ||
| + | *** NotI | ||
| + | |||
| + | ** pSB1C3_MPA (BBa_K494000) run on 2 % agarose gel | ||
| + | [[Image:TUM2010 101025 MPA.PNG|center|500 px]] | ||
| + | |||
| + | **--> clone 1 picked for submission: Looked better in reality! | ||
| + | |||
| + | ** BBa_K494001-BBa_K494006 run on 1.5 % agarose gel | ||
| + | [[Image:TUM2010 101025 backbones.PNG|center|500 px]] | ||
| + | |||
| + | ** pSB1C3_MPA (BBa_K494000) missed preps run on 2 % agarose gel | ||
| + | |||
| + | ===26.10.2010=== | ||
| + | * Waited for the FedEx man from 8:00-15:00. Called at 15:15. Said, oh, they forgot to pick it up! Sent somebody immediately who arrived at 15:45. Happyness --> Parts submitted, not our fault anymore :) Track: 871353522440 | ||
| + | |||
| + | ===27.10.2010=== | ||
| + | * Ran around to shoot weird ''E. coli'' pics | ||
| + | |||
| + | * Not in the lab anymore... FedEx delivered our parts! 9.14 am EDT, we recognized it at 16:22 MEST! More Happyness, now back to serious working maybe... | ||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | <br> | ||
| + | |||
| + | =Protocols= | ||
| + | |||
| + | ==Molecular Biology== | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | *PCR | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | |||
| + | <center>'''Taq Polymerase Hot Start '''</center> | ||
| + | |||
| + | '''PCR Pippeting plan: '''<br> | ||
| + | |||
| + | 1 µl template <br> | ||
| + | |||
| + | 1 µl dNTP 10 µM <br> | ||
| + | |||
| + | 1 µl G1004 (Primer) 10 µM<br> | ||
| + | |||
| + | 1 µl G1005 (Primer) 10 µM<br> | ||
| + | |||
| + | 5 µl 10x Taq-buffer (500 mM KCl, 100 mM Tris-HCl (pH 8.3), 15 mM MgCl<sub>2</sub>) | ||
| + | |||
| + | 0,2 µl Taq-Polymerase (add last) 5,000 U/ml<span style="background-color: rgb(255, 0, 0);"> | ||
| + | </span> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | 40.8 µl Water<br> | ||
| + | |||
| + | Final volume 50µl | ||
| + | |||
| + | '''<br>''' | ||
| + | |||
| + | '''Processing: '''(program saved as '''IGEMPCR ''')'''<br>''' | ||
| + | |||
| + | *preheating of PCR chamber to 94 °C | ||
| + | |||
| + | --> insert sample | ||
| + | |||
| + | *2 min at 94 °C | ||
| + | *loop 35x: | ||
| + | |||
| + | - 30 sat 94°C (according to IGEM protocols) | ||
| + | |||
| + | - 30 s at 56 °C | ||
| + | |||
| + | - 45s at 72°C | ||
| + | |||
| + | *7 min at 72°C | ||
| + | *stay at 4°C <br><br> | ||
| + | <center>'''colony PCR '''</center> | ||
| + | *Colony PCR | ||
| + | **pick colonies and resuspend them in 20 µl LB+Antibiotic (each) | ||
| + | **PCR of 2 µl of each sample, 2 µl as negative control (Program: ColonyPCR, modified), store remaining 18 µl for overnight cultures | ||
| + | **afterwards, mix 15 µl of each PCR product with 3 µl GLPn and load to Gel | ||
| + | **make overnight cultures of positive clones by adding the remaining 18 µl to 5 ml LB+AB | ||
| + | |||
| + | '''program:colonypcr ''' | ||
| + | |||
| + | *preheating of PCR chamber to 94 °C | ||
| + | |||
| + | --> insert sample | ||
| + | |||
| + | *5 min 30 sec at 94 °C | ||
| + | *loop 35x: | ||
| + | **30 sat 94°C (according to IGEM protocols) | ||
| + | **30 s at 58 °C | ||
| + | **60s at 72°C | ||
| + | *7 min at 72°C | ||
| + | *stay at 4°C | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | *DNA Purification | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | <center>'''PCR samples '''</center> | ||
| + | |||
| + | ''' ZYMO RESEARCH DNA Clean&Concentration Kit ''' | ||
| + | |||
| + | [http://www.zymoresearch.com/zrc/pdf/D4003i.pdf Protocol and Information]<br> | ||
| + | |||
| + | #In a 1.5 ml microcentrifuge tube, add 2-7 volumes of DNA Binding Buffer to each volume of DNA sample (see table below). Mix briefly by vortexing.<br><br> | ||
| + | |||
| + | {| cellspacing="1" cellpadding="1" border="1" align="center" width="80%" | ||
| + | |- | ||
| + | | Application | ||
| + | | DNA Binding Buffer : Sample | ||
| + | | Example | ||
| + | |- | ||
| + | | Plasmid, genomic DNA (>2 kb) | ||
| + | | 2 : 1 | ||
| + | | 200 μl : 100 μl | ||
| + | |- | ||
| + | | PCR, cDNA, DNA fragment | ||
| + | | 5 : 1 | ||
| + | | 500 μl : 100 μl | ||
| + | |- | ||
| + | | ssDNA (e.g., M13 phage) | ||
| + | | 7 : 1 | ||
| + | | 700 μl : 100 μl | ||
| + | |} | ||
| + | |||
| + | #Transfer mixture to a provided Zymo-Spin™ Column1 in a Collection Tube.<br> | ||
| + | #Centrifuge at ≥10,000 x g for 30 seconds. Discard the flow-through.<br> | ||
| + | #Add 200 μl Wash Buffer to the column. Centrifuge at ≥10,000 x g for 30 seconds. Repeat wash step.<br> | ||
| + | #Add ≥6 μl water2,3 directly to the column matrix. Transfer the column to a 1.5 ml microcentrifuge tube and centrifuge at ≥10,000 x g for 30 seconds to elute the DNA.<br>Ultra-pure DNA in water is now ready for use.<br> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | ''' QIAquick purification Kit <br> ''' | ||
| + | |||
| + | [http://www1.qiagen.com/literature/render.aspx?id=103715 Handbook] | ||
| + | |||
| + | Procedure<br> 1. Add 5 volumes of Buffer PB to 1 volume of the PCR sample and mix. It is not necessary to remove mineral oil or kerosene. For example, add 500 μl of Buffer PB to 100 μl PCR sample (not including oil).<br> 2. If pH indicator I has beein added to Buffer PB, check that the color of the mixture is yellow. If the color of the mixture is orange or violet, add 10 μl of 3 M sodium acetate, pH 5.0, and mix. The color of the mixture will turn to yellow.<br> 3. Place a QIAquick spin column in a provided 2 ml collection tube. <br>4. To bind DNA, apply the sample to the QIAquick column and centrifuge for 30–60 s. '''We changed it to 3 min @ 6000rpm ! '''<br>5. Discard flow-through. Place the QIAquick column back into the same tube. Collection tubes are re-used to reduce plastic waste.<br> 6. To wash, add 0.75 ml Buffer PE to the QIAquick column and centrifuge for 30–60 s.<br> 7. Discard flow-through and place the QIAquick column back in the same tube. Centrifuge the column for an additional 1 min.'''repeat!'''<br> IMPORTANT: Residual ethanol from Buffer PE will not be completely removed unless the flow-through is discarded before this additional centrifugation.<br> 8. Place QIAquick column in a clean 1.5 ml microcentrifuge tube.<br> 9. To elute DNA, add 50 μl Buffer EB (10 mM Tris·Cl, pH 8.5) or water (pH 7.0–8.5) to the center of the QIAquick membrane and centrifuge the column for 1 min. Alternatively, for increased DNA concentration, add 30 μl elution buffer to the center of the QIAquick membrane, let the column stand for 1 min, and then centrifuge.<br> IMPORTANT: Ensure that the elution buffer is dispensed directly onto the QIAquick membrane for complete elution of bound DNA. The average eluate volume is 48 μl from 50 μl elution buffer volume, and 28 μl from 30 μl elution buffer. Elution efficiency is dependent on pH. The maximum elution efficiency is achieved between pH 7.0 and 8.5. When using water, make sure that the pH value is within this range, and store DNA at –20°C as DNA may degrade in the absence of a buffering agent. The purified DNA can also be eluted in TE buffer (10 mM Tris·Cl, 1 mM EDTA, pH 8.0), but the EDTA may inhibit subsequent enzymatic reactions.<br> 10. If the purified DNA is to be analyzed on a gel, add 1 volume of Loading Dye to 5 volumes of purified DNA. Mix the solution by pipetting up and down before loading the gel.<br> | ||
| + | <center>''' Gel samples<br> '''</center> | ||
| + | |||
| + | ''' ZYMO RESEARCH Gel DNA Recovery Kit ''' | ||
| + | [http://www.acgtinc.com/PDF_files/Sample%20Prepation_ACGT/Zymoclean%20Gel%20DNA%20Recovery%20Kit_Zymo%20Research.pdf Product informartion] | ||
| + | |||
| + | '''Protocol'''<br> | ||
| + | |||
| + | #Excise the DNA fragment1 from the agarose gel using a razor blade or scalpel and transfer it to a 1.5 ml microcentrifuge tube. | ||
| + | #Add 3 volumes of ADB to each volume of agarose excised from the gel (e.g. for 100 μl (mg) of agarose gel slice add 300 μl of ADB). | ||
| + | #Incubate at 37-55 °C for 5-10 minutes until the gel slice is completely dissolved2. For DNA fragments >8 kb, following the incubation step, add one additional volume (equal to that of the gel slice) of water to the mixture for better DNA recovery (e.g. 100 μl agarose, 300 μl ADB and 100 μl water). | ||
| + | #Transfer the melted agarose solution to a Zymo-SpinTM I Column in a Collection Tube. | ||
| + | #Centrifuge at ≥10,000 x g for 30-60 seconds. Discard the flow-through. | ||
| + | #Add 200 μl of Wash Buffer to the column and centrifuge at ≥10,000 x g for 30 seconds. Discard the flow-through. Repeat the wash step. | ||
| + | #Add ≥6 μl of water3,4 directly to the column matrix. Place column into a 1.5 ml tube and centrifuge ≥10,000 x g for 30-60 seconds to elute DNA.<br>Ultra-pure DNA in water is now ready for use.<br> | ||
| + | |||
| + | <center>''' Miniprep '''</center> | ||
| + | |||
| + | '''Protocol:''' | ||
| + | |||
| + | # Add 600 μl of bacterial culture grown in LB medium to a 1.5 ml microcentrifuge tube. | ||
| + | # Add 100 μl of 7X Lysis Buffer (Blue)1 and mix by inverting the tube 4-6 times. Proceed to step 3 within 2 minutes. After addition of 7X Lysis Buffer the solution should change from opaque to clear blue, indicating complete lysis. | ||
| + | # Add 350 μl of cold Neutralization Buffer (Yellow)2 and mix thoroughly. The sample will turn yellow when the neutralization is complete and a yellowish precipitate will form. Invert the sample an additional 2-3 times to ensure complete neutralization. | ||
| + | # Centrifuge at 11,000 – 16,000 x g for 2-4 minutes. | ||
| + | # Transfer the supernatant (~900 μl) into the provided Zymo-Spin™ IIN column. Avoid disturbing the cell debris pellet. | ||
| + | # Place the column into a Collection Tube and centrifuge for 15 seconds. | ||
| + | # Discard the flow-through and place the column back into the same Collection Tube. | ||
| + | # Add 200 μl of Endo-Wash Buffer to the column. Centrifuge for 15 seconds. It is not necessary to empty the collection tube. | ||
| + | # Add 400 μl of Zyppy™ Wash Buffer2 to the column. Centrifuge for 30 seconds. | ||
| + | # Transfer the column into a clean 1.5 ml microcentrifuge tube then add 30 μl of Zyppy™ Elution Buffer3 directly to the column matrix and let stand for one minute at room temperature. | ||
| + | # Centrifuge for 15 seconds to elute the plasmid DNA. | ||
| + | |||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | *Digestion | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | <center>''' Restriction Digest '''</center> | ||
| + | |||
| + | {| width="60%" cellspacing="1" cellpadding="1" border="1" | ||
| + | |- | ||
| + | | Enzyme<br> | ||
| + | | 10 units is sufficient, generally 1µl is used | ||
| + | |- | ||
| + | | DNA | ||
| + | | 1 µg | ||
| + | |- | ||
| + | | 10X NEBuffer<br> | ||
| + | | 5 µl (1X) | ||
| + | |- | ||
| + | | BSA | ||
| + | | Add to a final concentration of 100 µg/ml (1X) if necessary | ||
| + | |- | ||
| + | | Total Reaction Volume | ||
| + | | 50 µl | ||
| + | |- | ||
| + | | Incubation Time | ||
| + | | 1 - 1.5 hour<br> | ||
| + | |- | ||
| + | | Incubation Temperature Enzyme dependent | ||
| + | | | ||
| + | XbaI, SpeI, PstI, SpeI : 37 °C | ||
| + | |||
| + | |} | ||
| + | |||
| + | [http://www.neb.com/nebecomm/tech_reference/restriction_enzymes/buffer_activity_restriction_enzymes.asp activity of restriction enzymes in NEB buffers] <br> | ||
| + | |||
| + | ''' Biobrick standard <br> ''' | ||
| + | |||
| + | [http://openwetware.org/wiki/Restriction_digest Protocols for IGEM standard digestion] | ||
| + | <br> | ||
| + | <center>'''Dephosphorylation'''</center> | ||
| + | using Antarctic Phosphatase | ||
| + | #Add 1/10 volume of 10X Antarctic Phosphatase Reaction Buffer to 1-5 µg of DNA cut with any restriction endonuclease in any buffer. | ||
| + | #Add 1 µl of Antarctic Phosphatase (5 units) and mix. | ||
| + | #Incubate for 15 minutes at 37°C for 5´ extensions or blunt-ends, 60 minutes for 3´ extensions. | ||
| + | #Heat inactivate (or as required to inactivate the restriction enzyme) for 5 minutes at 65°C. | ||
| + | #Proceed with ligation. | ||
| + | [http://www.neb.com/nebecomm/products/protocol76.asp from NEB] | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | *Ligation | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | ''' Using T4 Ligase, New England Labs ''' | ||
| + | *1 µl T4 Ligase <span style="color: rgb(255, 102, 0);">(10.000 U)</span> | ||
| + | *50 ng plasmid | ||
| + | *3x mol(plasmid) insert | ||
| + | *2 µl T4 Ligase 10x buffer | ||
| + | *add H<sub>2</sub>O to reach final volume of 20 µl<br> | ||
| + | |||
| + | *incubation at 22°C for 1 h | ||
| + | *storing at 16 °C for 40 min<br> | ||
| + | |||
| + | |||
| + | <u>'''Biobrick Standard'''</u><br> | ||
| + | |||
| + | [http://parts2.mit.edu/wiki/index.php/Standard_Assembly Standard BioBrick assembly]<br> | ||
| + | |||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | *Transformation | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | |||
| + | '''At Woehlke's S1-Lab !!!'''<br> | ||
| + | |||
| + | #Thaw competent cells on Ice | ||
| + | #Add DNA, pipette gently to mix | ||
| + | #Let sit for 30 minutes on ice | ||
| + | #Incubate cells for 45 seconds at 42°C | ||
| + | #Incubate cells on ice for 2 min | ||
| + | #Add 1 ml LB0 | ||
| + | #Incubate for 1 hour at 37oC on shaker | ||
| + | #Spread 100-300 μl onto a plate made with appropriate antibiotic. | ||
| + | #Grow overnight at 37 °C. | ||
| + | #Save the rest of the transformants in liquid culture at 4 °C | ||
| + | |||
| + | modified from [http://openwetware.org/wiki/Transforming_chemically_competent_cells open wetware] | ||
| + | |||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | *Gel electrophoresis | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | <center>'''Agarose Gels '''</center> | ||
| + | usual volume needed: 80 ml | ||
| + | [http://www.promega.com/enotes/faqspeak/fq0065.htm Optimum resolution according to NEB]<br> | ||
| + | [http://www.cwcboe.org/19992051412432550/lib/19992051412432550/Molecular%20Biology/Gel%20electrophoresis/Lab_manual_8_gel_elect.pdf further information on optimizing gel electrophoresis, e.g. recommanded voltage per cm2 gel] | ||
| + | |||
| + | '''stain''' | ||
| + | |||
| + | *SybrGold ([http://www.invitrogen.com/site/us/en/home/References/Molecular-Probes-The-Handbook/Nucleic-Acid-Detection-and-Genomics-Technology/Nucleic-Acid-Detection-and-Quantitation-in-Electrophoretic-Gels-and-Capillaries.html invitrogen]) | ||
| + | **Cover Gel with 1x TAE | ||
| + | **Add SybrGold to a 1:10000 dilution | ||
| + | **cover with aluminium foil (light sensitive) | ||
| + | **shake&incubate 20 min (for 2% Agarose Gels at least 45 min!) | ||
| + | <br> | ||
| + | *SybrSafe | ||
| + | **used just like SybrGold | ||
| + | |||
| + | ''' standards<br> ''' | ||
| + | |||
| + | |||
| + | |||
| + | <br> | ||
| + | |||
| + | {| width="600" height="386" cellspacing="1" cellpadding="1" border="1" align="center" style="" | ||
| + | |- | ||
| + | | [[Image:TUM2010_Dna lmw.gif]] <br> | ||
| + | | [[Image:TUM2010_N3232_fig1_v1_000034.gif]]<br> | ||
| + | | [[Image:TUM2010_N3200_fig1_v1_000036.gif]]<br> | ||
| + | |- | ||
| + | | low molecular weight (NEB)<br> | ||
| + | | 1 kb standard (NEB)<br> | ||
| + | | 2-log standard (NEB)<br> | ||
| + | |} | ||
| + | |||
| + | <br> | ||
| + | <center>'''Polyacrylamide Gels'''</center> | ||
| + | '''Preparation of Gels''' | ||
| + | |||
| + | Recipe for denaturing gels: | ||
| + | {| cellspacing="1" cellpadding="1" border="1" align="center" width="80%" | ||
| + | |- | ||
| + | | Gel type | ||
| + | | 1 big gel | ||
| + | | 2 big gels | ||
| + | | 1 small gel | ||
| + | | 2 small gels | ||
| + | |- | ||
| + | | Urea | ||
| + | | 28.8 g | ||
| + | | 57.6 g | ||
| + | | x | ||
| + | | x | ||
| + | |- | ||
| + | | Acrylamide 40% | ||
| + | | 22.5 ml | ||
| + | | 45 ml | ||
| + | | x | ||
| + | | x | ||
| + | |- | ||
| + | | Buffer 10x | ||
| + | | 6 ml | ||
| + | | 12 ml | ||
| + | | x | ||
| + | | x | ||
| + | |- | ||
| + | | End volume (reach by adding water) | ||
| + | | 60 ml | ||
| + | | 120 ml | ||
| + | | x | ||
| + | | x | ||
| + | |- | ||
| + | | APS | ||
| + | | 600 µl | ||
| + | | 1200 µl | ||
| + | | x | ||
| + | | x | ||
| + | |- | ||
| + | | TEMED | ||
| + | | 60 µl | ||
| + | | 120 µl | ||
| + | | x | ||
| + | | x | ||
| + | |- | ||
| + | |} | ||
| + | <br> | ||
| + | *Dissolve Urea in Acrylamide-buffer mixture (use Ultrasound bath), this may take more than an hour! | ||
| + | *Tighten the Gel chamber | ||
| + | *add water to desired end volume | ||
| + | *Add APS, then TEMED, mix | ||
| + | *Pipette mixture into gel chamber | ||
| + | *Add desired comb | ||
| + | *let gel polymerize overnight; add buffer in the evening | ||
| + | |||
| + | '''Running of Gels''' | ||
| + | |||
| + | mix samples 1:1 with formamide loading dye (stored @ -20°C) | ||
| + | carefully remove comb | ||
| + | blow air into pockets with a 50 µl syringe | ||
| + | fill samples into pockets | ||
| + | run the gel | ||
| + | (usually about 200 V) | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | |||
| + | ==''In vivo'' Measurement== | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | ===Bacterial Cell Growth=== | ||
| + | Bacteria from over night cultures were diluted 1:50 into 20 ml culture in LBamp and incubated at 37 °C. Upon OD<sub>600</sub> of 0.7-0.8 the cultures were induced with 0.4% Arabinose and 0.4% Arabinose + 1mM IPTG, respectively. Subsequently Cultures were incubated at 25°C for at least 12 h. | ||
| + | |||
| + | ===Fluorescence Measurement=== | ||
| + | Cell samples for the fluorescence measurement were diluted to OD<sub>600</sub>=0.03 and analyzed in a JASCO fluorimeter. eGFP excitation wavelength was set to 501 nm and mCherry fluorescence was measured with an excitation at 587 nm. Standard parameters for the fluorimeter included scanning speed of 100 nm/ min and data points every 0.2 nm as well as medium detector sensivity. The cuvette holder was temperated to 25 °C. | ||
| + | The resulting spectra were corrected for instrumental wavelength dependencies and quantum yield of the fluorescent proteins. A pure LBamp spectrum was subtracted and the corrected spectra were normalized using eGFP fluorescence as reference. | ||
| + | |||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | |||
| + | ==''In vitro'' Translation== | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | The [http://www.promega.com/catalog/catalogproducts.aspx?categoryname=productleaf_335&ckt=1 Promega Kit] is used according to the provided protocols. Further Information about this Kit can be found in the [http://partsregistry.org/Cell-free_chassis/Commercial_E._coli_S30 Parts Registry]. | ||
| + | |||
| + | Fluorescence kinetics are recorded for at least 3 hours, settings are applied as in the in vitro measurement. | ||
| + | |||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | |||
| + | ==''In vitro'' Transcription== | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | ===Buffers=== | ||
| + | Three different buffers were used for ''in vitro'' transcription experiments: | ||
| + | * For Epicentre E. coli RNA Polymerase the recommended buffer was used | ||
| + | * For T7 RNA Polymerase experiments two buffers were tested: | ||
| + | **T7 RPO buffer as recommended and used by members of the Simmel group | ||
| + | **T7 RPO "paper buffer", as used in the paper of XXX | ||
| + | Of all buffers 2x concentrated stocks were prepared. Malachite green was usually added to the buffer stocks. | ||
| + | {| cellspacing="1" cellpadding="1" border="1" align="center" width="80%" | ||
| + | |- | ||
| + | | | ||
| + | | E. coli RPO buffer | ||
| + | | T7 RPO buffer | ||
| + | | T7 RPO buffer "paper" | ||
| + | |- | ||
| + | | Tris | ||
| + | | 40 mM | ||
| + | | 40 mM | ||
| + | | 40 mM | ||
| + | |- | ||
| + | | pH | ||
| + | | 7.5 | ||
| + | | 7.1 | ||
| + | | 7.9 | ||
| + | |- | ||
| + | | MgCl2 | ||
| + | | 10 mM | ||
| + | | 40 mM | ||
| + | | 6 mM | ||
| + | |- | ||
| + | | KCl | ||
| + | | 150 mM | ||
| + | | / | ||
| + | | 100 mM | ||
| + | |- | ||
| + | | Triton X-100 | ||
| + | | 0.01% | ||
| + | | / | ||
| + | | / | ||
| + | |- | ||
| + | |} | ||
| + | ===Sample Preparation=== | ||
| + | Different concentrations were tested for malachite green and DNA templates. Components of a standard experiment are listed in the table below. | ||
| + | |||
| + | {| cellspacing="1" cellpadding="1" border="1" align="center" width="80%" | ||
| + | |- | ||
| + | | | ||
| + | | E. coli RPO buffer | ||
| + | | T7 RPO buffer | ||
| + | | T7 RPO buffer "paper" | ||
| + | |- | ||
| + | | buffer | ||
| + | | 1x | ||
| + | | 1x | ||
| + | | 1x | ||
| + | |- | ||
| + | | DTT | ||
| + | | 10 mM | ||
| + | | 10 mM | ||
| + | | 10 mM | ||
| + | |- | ||
| + | | Malachite green | ||
| + | | 5-10 µM | ||
| + | | 5-10 µM | ||
| + | | 5-10 µM | ||
| + | |- | ||
| + | | NTP's | ||
| + | | 1 mM | ||
| + | | 4 mM | ||
| + | | 0.8 mM | ||
| + | |- | ||
| + | | RPO | ||
| + | | 2 U | ||
| + | | 125 U | ||
| + | | 125 U | ||
| + | |- | ||
| + | |} | ||
| + | In each run up to 4 samples are measured simultaneously. Components that are the same in each of the 4 samples (buffer, DTT, NTPs, RPO, Water) are prepared as a 4.1x MasterMix in a loBind tube and split to the 4 cuvettes. Final Volume of each sample is 100 µl. | ||
| + | ===Cary Eclipse=== | ||
| + | For fluorescence measurements, a Cary Eclipse Spectrofluorimeter with a Multicellholder (4 cells) is used. Kinetics are recorded at 37° C. Excitation wavelength is 630 nm, emission is followed at 650 nm and 655 nm. After each kinetics measurement, spectra are to be recorded. | ||
| + | |||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | |||
| + | =References= | ||
| + | <html><a name="ref1"></a></html>[1] http://www.promega.com/catalog/catalogproducts.aspx?categoryname=productleaf_335&ckt=1 | ||
| + | <html><a name="ref2"></a></html>[2] Zubay, G. (1980) Meth. Enzymol. 65, 856–77, Zubay, G. (1973) Ann. Rev. Genet. 7, 267–87. | ||
<!-- ############## WIKI-PAGE STOPS HERE ############## --> | <!-- ############## WIKI-PAGE STOPS HERE ############## --> | ||
Latest revision as of 12:04, 22 November 2010
|
|
Experiment DesignIn this section we do not only want to present the experiments and results we gained but also to encourage you to evaluate your own switch based on the protocols and general procedure on how to evaluate basic parameters of a switch. In theory, every terminator can be turned into a switch with minor modifications and the right signals which are based on individual applications. While the principle of how to turn a terminator into a switch is explained in detail here, experimental setups and protocols are provided in the following. Due to time and equipment limitations we could not perform all the experiments we planned but next to the hope that another iGEM team might proceed with our project we would also like to encourage you to design and test some basic switches on which you can base a complete, tightly regulated network.
The complexity of our experimental setups vary, since we planned to characterize an individual switch with one exemplary signal on all relevant levels: Starting from the most general, complicated but also relevant level, in vivo measurements we approached to testing different switches on each smaller scale: We developed setups for in vitro translation which can be done without much effort following the in vitro measurements and also provide detailed description of in vitro transcription verification providing an inside to the molecular functionality of our basic idea. We do not see the methods we used here as the gold standard for bioLOGICS evaluation and encourage you to include your own ideas as well as check in our outlook section where we suggest experiments we could not do during the limited iGEM 2010 time. Together with our Biobrick submissions this year, we offer a complete set for switch evaluation on all cellular levels.
In vivo MeasurementsIn vivo measurements have the highest complexity compared to any other experimental set-up. Different parameters and circumstances deriving from both the cellular environment as well as technical considerations like scatter have to be taken into account. Nevertheless, the measurements are essential, as our switches should finally work inside cells to fulfill our vision of an intracellular logic network. This year's submitted Biobricks provide you with a basic kit of plasmids which allow a quick beginning of the measurements.
DesignFor the measurements in vivo we decided to use an expression cassette consisting of Green Fluorescent Protein (GFP) coding sequence upstream of the switch and another fluorescent protein coding sequence downstream of it. Both protein coding sequence share the same ribosome binding site allowing the usage of the GFP as an internal control in measurements. Since the spectra should not overlap and to avoid FRET as well as an pure overlap of the spectra, we settled on the usage of red fluorescent protein variants, namely mRFP1 in the first try and mCherry in an modified variant of the pSB1A10 vector.
The GFP internal control carries the advantage that errors in the measurement set can be detected easily. Lack of arabinose or promoter insensitivity can be recognized as well as problems with the fluorescence measurement itself. Plus, it allows normalizing measurements to compare different preparations in relation to each other.
Construction and CloningIn a first try we cloned a measuring construct based on pSB1A10. The resulting plasmid, nicknamed pMonsterplasmid due to its size was tested in the fluorescent measurements described below. Unfortunately after two months of cloning we had to recognize that the plasmid in use did not work for us (see also pSB1A10 Falsification).
Measurements based on submitted BiobricksThe Biobricks BBa_K494001-BBa_K494006 are constructed for easy design of a switch-evaluation system. Detailed information can be found here. Switch evaluation in vivoTo evaluate the switching efficiency, output with and without signal needs to be monitored. In this case, GFP fluorescence (internal control) will always appear upon arabinose induction, while RFP/mCherry fluorescence is only present upon binding of a signal and occurring antitermination.
In vitro TranslationTo go more into detail, the next complexity level is to study the effect of switches on a translational level. In vitro measurements with E. coli lysate make the fluorescence signals independent of cell growth and physical or biological factors like cell density or growth stadium.
DesignSince the same constructs can be used both for in vivo and in vitro translation, no additional cloning effort is needed. This implements, that the Biobricks we provided this year, can again be used as the groundwork for constructing vectors for measurements. MeasurementsWe used the cell-free E. coli S30 extract system for circular DNA provided by Promega[1], which is prepared by modifications of the Method Zubay et al.[2]. The characterization of the kit can be obtained from the [http://partsregistry.org/Cell-free_chassis/Commercial_E._coli_S30 Parts Registry]. In vitro TranscriptionTo monitor transcription termination and antitermination on a the molecular level, in vitro transcription of individual switches and their response to signals offer an elegant way for fast and easy prove of principle. Most side effects occurring in a complex environment given in a cell or a cell lysate do not arise here. Another major advantage of in vitro transcription experiments is the possibility to test many signals for one switch to optimize antitermination efficiency and binding specificity without much cloning work. Data gained by in vitro transcription experiments can be used to improve switches and signals for in vivo usage.
Since we are working on a totally new principle of transcriptional control, we used this approach beside the above mentioned advantages for easy variation of different variables like the length of the core unit and the switch to signal ratio. T7 RNA polymeraseThe T7 RNA polymerase is known for satisfying RNA yields together with easy handling. In our approach we had PCR amplified, double stranded switches with an malachitegreen binding aptamer following after a T7 terminator which was constructed to function as a switch. Different signals were tested varying in length of the specificity site and the triggering unit.
E. coli RNA polymeraseIn comparison to the T7 RNA Polymerase the E. coli RNA Polymerase requires slightly more sophisticated proceedings when it comes to the design of switches and handling of the enzyme. The biggest in our case was to store it properly since the only -80°C fridge was in another building, so make sure you have a big supply of dry ice ready if you encounter the same problem. Denaturing Polyacrylamide gel electrophoresisPolyacrylamide gel electrophoresis (PAGE) was used for evaluation of termination and switching efficiency. Gels containing 15 % acrylamide and 6 M urea were used for separation of terminated and readthrough RNAs. The same constructs as designed for the malachite green binding aptamer were used.
Malachite green assayIn this year's DNA submission we contribute the malachite green binding aptamer which can be used as a transcription reporter in in vitro transcription experiments. Malachite green binding can be used to follow RNA transcription over time, a rise in the fluorescence is then detectable. Fluorescence marker of specific RNA structures are still rare, so the malachite green binding aptamer provides one of the only possibilities to continuously monitor transcription reactions. In comparison to PAGE, kinetics can be taken, while with PAGE only end point estimations can be made. This makes the malachite green binding aptamer a valuable tool to study in vitro transcription in general and the principles underlying our switch in principle.
Lab BookExplanationsIn the following we present an overview regarding our work in the lab. For easier understanding we summarized the work of each week using colored boxes. To get a better overview we used the following color code for the boxes:
Chronological Lab Bookin vivo constructs
08.04.2010Flo & Philipp PCR
09.04.2010Philipp & Flo Gel of PCR products from 08.04.2010
in vivo constructs
15.04.2010Philipp & Flo
16.04.2010Philipp & Flo
--> worked for R0011, not for B0014
* approximated from the amount used in the digestion beforeClose in vivo constructs
19.04.2010
* approximated from the amount used in the digestion before 20.04.2010
21.04.2010
File:TUM2010 100421beschriftet.gif??
--> worked!!!!!
22.04.2010
23.04.2010
in vivo constructs
26.04.2010
27.04.2010
Many colonies with pSB1K3-B0014, not one with pSB1K3-Sig-B0014
28.04.2010
29.04.2010
30.04.2010PCR R0011-HisSig and R0011-TrpSig --> 13 ng/µl x 20 µl Close
in vivo constructs
04.05.2010
--> each in 600 µl LB+Carbenicillin (=Ampicillin) @37°C 05.05.2010
06.05.2010
07.05.2010
in vivo constructs
10.05.2010
Prefix: 20 bp; Suffix: 21 bp; X-S-scar: 6 bp --> it looks as if R0011 is ligated to B0014, which makes the whole construct wothless. The R0011-HisSig control looks more like R0011 alone as well. 11.05.2010
12.05.2010
File:TUM2010 100512beschriftet.png
14.05.2010
File:TUM2010 100514beschriftet.png
in vivo constructs
17.05.2010
18.05.2010
Samples: LMW|R0011|HisSig|TrpSig|HisSig E/S-Dig|TrpSig E/S-Dig|B0014|LMW2|Colony PCR His1|His2|Trp1|Trp2|control|HisTerm|TrpTerm|HisTerm E/P-Dig|TrpTerm E/P-Dig
Important Mistake! See below Gel!
(*R0011 and B0014 look normal
--> are all of our sequences just wrong??? What are we going to do? Order everything new?) 19.05.2010
File:TUM2010 100519beschriftet.png600px 20.05.2010
21.05.2010
in vivo constructs
25.05.2010
26.05.2010
27.05.2010
28.05.2010
in vivo constructs
31.05.2010
01.06.2010
02.06.2010
JobNr. Barcode Last change Date/Time Last message / Files 6549287 AE2739 02.06.2010 / 13:51:12 HisSig 1-3-forward G1004 We just received your order. Many thanks. 6549288 AE2738 02.06.2010 / 13:51:12 HisSig 3-1-forward G1004 We just received your order. Many thanks. 6549289 AE2737 02.06.2010 / 13:51:12 TrpSig DL4-1-forward G1004 We just received your order. Many thanks. 6549290 AE2736 02.06.2010 / 13:51:12 TrpSig DL4-3-forward G1004 We just received your order. Many thanks. 6549291 AE2735 02.06.2010 / 13:51:12 HisTerm-forward G1004 We just received your order. Many thanks. 6549292 AE2734 02.06.2010 / 13:51:12 TrpTerm-forward G1004 We just received your order. Many thanks.
in vivo constructs
07.06.2010
08.06.2010
09.06.2010
10.06.2010
11.06.2010
in vivo constructs Promega Kit
14.06.2010
15.06.2010
16.06.2010Fluoresence measurements using in vitro kit
Cell culture 5 ml culture for
17.06.2010 Cloning
18.06.2010cloning
2 % broad range agarose, 1 h 120 V
in vitro measurements
f$%&&§ s%§$! Again, nothing worked! Although we saw an increasing "GFP signal" comparable to 16.06.10, taking spectra suggested we DON'T see significant GFP-production! We used new water for preparing the samples, cleaned cuvettes with "new water", used other DNA-samples etc. Somehow, it seems as if we don't express GFP (we compared Christoph's results! We should see a really significant spectrum!
in vivo constructs First steps Promega Kit
22.06.2010Transformation
Liquid culture
23.06.2010
0.2% 24.06.2010
25.06.2010Plasmid purification and sequencing
Fluorescence Measurements
in vivo constructs Testing pSB1A10
28.06.2010
29.06.2010Closein vivo constructs Invitrogen Kit
01.07.2010
02.07.2010
Testing pSB1A10
05.07.2010
06.07.2010
Week16 The Era of Exams No Lab work this week, everybody is busy studying for their exams... CloseWeek17 The Era of Exams No Lab work this week, everybody is busy studying for their exams... Close
Week18 The Era of Exams No Lab work this week, everybody is busy studying for their exams... Close
Week19 The Era of Exams No Lab work this week, everybody is busy studying for their exams... CloseConstruction of new Measurement Plasmid
09.08.2010
10.08.2010
(=> no colonies the next day)
(=> pSB1A10-TrpTerm and pSB1A10-HisTerm did not grow until next day)
11.08.2010
12.08.2010
Caution: ran out of gas => not steril? 13.08.2010
Construction of new Measurement Plasmid
16.08.2010
17.08.2010
PROBLEM: mCherry has a SgrA1-cleavage site! These constructs cannot be used. Starting all over, cloning the linker sequence first... 18.08.2010
19.08.2010
20.08.2010
Construction of new Measurement Plasmid
23.08.2010
24.08.2010
Faint bands at the correct length can be guessed.
25.08.2010
26.08.2010
27.08.2010
for sequencing
Construction of new Measurement Plasmid Testing new Measurement Plasmid
30.08.2010
Gel 1: => samples named "1x" to "8x" for pSB1A2-R0011-BB1006Sig-B0014 colonies => samples named "9x" to "12x" for pSB1A10mod-TrpTerm-mCherry-TrpSig colonies Gel 2: => samples named "13x" to "16x" for pSB1A10mod-TrpTerm-mCherry-TrpSig colonies => samples named "17x" to "24x" for pSB1A10mod-HisTerm-mCherry-HisSig colonies Interpretation for Gel1 and Gel2: Ligation worked for the samples 1x-6x (pSB1A2-R0011-BB1006Sig-B0014), 9x-16x (pSB1A10mod-TrpTerm-mCherry-TrpSig), 18x-24x (pSB1A10mod-HisTerm-mCherry-HisSig)
31.08.2010
01.09.2010
02.09.2010Starting the cloning of pBAD (BioBrick I13453) downstream of GFP
03.09.2010
in vivo constructs Testing HisTrp
06.09.2010
07.09.2010
08.09.2010
-->OD600 0.05 reasonable for our cell measurements. Positive control works fine after 24 h induction.
09.09.2010
10.09.2010
His-Switch DOES NOT seem to work!!! Measurements of Trp-Switch look weird, since Ara-induced culture showed strong mCherry signal than Ara+IPTG-induced culture. Maybe growing condition were not sufficient. We will use 50 ml flask next time! Close
Cloning HisTerm/TrpTerm Ordering Aptamer New Measurement Plasmid
13.09.2010
14.09.2010
15.09.2010
16.09.2010
17.09.2010
Cloning of Malachit green constructs HisTerm/TrpTerm
20.09.2010
21.09.2010
22.09.2010
23.09.2010
24.09.2010
T7 Switch
27.09.2010
01.10.2010T7-Measurments
=> Switches do look quite good. However DTT was not the same in all samples.
T7 Switch
04.10.2010
05.10.2010
PCR T7 switch/positive signal
06.10.2010
07.10.2010
08.10.2010Close
T7 Switch
11.10.2010
12.10.2010
13.10.2010
14.10.2010
15.10.2010
17.10.2010
Cloning of Parts into pSB1C3 T7 & E. coli
18.10.2010
--> Favourite one ever so far!
19.10.2010
20.10.2010
21.10.2010
22.10.2010
23.10.2010
600px Only the positive control without Signal increases!
24.10.2010
Cloning of Parts into pSB1C3 T7 & E. coli
The Week of Wiki freeze! 25.10.2010
26.10.2010
27.10.2010
ProtocolsMolecular Biology
PCR Pippeting plan: 1 µl template 1 µl dNTP 10 µM 1 µl G1004 (Primer) 10 µM 1 µl G1005 (Primer) 10 µM 5 µl 10x Taq-buffer (500 mM KCl, 100 mM Tris-HCl (pH 8.3), 15 mM MgCl2) 0,2 µl Taq-Polymerase (add last) 5,000 U/ml
40.8 µl Water Final volume 50µl
Processing: (program saved as IGEMPCR )
--> insert sample
- 30 sat 94°C (according to IGEM protocols) - 30 s at 56 °C - 45s at 72°C
program:colonypcr
--> insert sample
ZYMO RESEARCH DNA Clean&Concentration Kit [http://www.zymoresearch.com/zrc/pdf/D4003i.pdf Protocol and Information]
QIAquick purification Kit [http://www1.qiagen.com/literature/render.aspx?id=103715 Handbook] Procedure ZYMO RESEARCH Gel DNA Recovery Kit [http://www.acgtinc.com/PDF_files/Sample%20Prepation_ACGT/Zymoclean%20Gel%20DNA%20Recovery%20Kit_Zymo%20Research.pdf Product informartion] Protocol
Protocol:
[http://www.neb.com/nebecomm/tech_reference/restriction_enzymes/buffer_activity_restriction_enzymes.asp activity of restriction enzymes in NEB buffers] Biobrick standard [http://openwetware.org/wiki/Restriction_digest Protocols for IGEM standard digestion]
using Antarctic Phosphatase
[http://www.neb.com/nebecomm/products/protocol76.asp from NEB] Close
Using T4 Ligase, New England Labs
[http://parts2.mit.edu/wiki/index.php/Standard_Assembly Standard BioBrick assembly]
At Woehlke's S1-Lab !!!
modified from [http://openwetware.org/wiki/Transforming_chemically_competent_cells open wetware] Close
usual volume needed: 80 ml
[http://www.promega.com/enotes/faqspeak/fq0065.htm Optimum resolution according to NEB] stain
standards
Preparation of Gels Recipe for denaturing gels:
Running of Gels mix samples 1:1 with formamide loading dye (stored @ -20°C) carefully remove comb blow air into pockets with a 50 µl syringe fill samples into pockets run the gel (usually about 200 V) CloseClose In vivo Measurement
Bacterial Cell GrowthBacteria from over night cultures were diluted 1:50 into 20 ml culture in LBamp and incubated at 37 °C. Upon OD600 of 0.7-0.8 the cultures were induced with 0.4% Arabinose and 0.4% Arabinose + 1mM IPTG, respectively. Subsequently Cultures were incubated at 25°C for at least 12 h. Fluorescence MeasurementCell samples for the fluorescence measurement were diluted to OD600=0.03 and analyzed in a JASCO fluorimeter. eGFP excitation wavelength was set to 501 nm and mCherry fluorescence was measured with an excitation at 587 nm. Standard parameters for the fluorimeter included scanning speed of 100 nm/ min and data points every 0.2 nm as well as medium detector sensivity. The cuvette holder was temperated to 25 °C. The resulting spectra were corrected for instrumental wavelength dependencies and quantum yield of the fluorescent proteins. A pure LBamp spectrum was subtracted and the corrected spectra were normalized using eGFP fluorescence as reference. CloseIn vitro Translation
The [http://www.promega.com/catalog/catalogproducts.aspx?categoryname=productleaf_335&ckt=1 Promega Kit] is used according to the provided protocols. Further Information about this Kit can be found in the [http://partsregistry.org/Cell-free_chassis/Commercial_E._coli_S30 Parts Registry]. Fluorescence kinetics are recorded for at least 3 hours, settings are applied as in the in vitro measurement. CloseIn vitro Transcription
BuffersThree different buffers were used for in vitro transcription experiments:
Of all buffers 2x concentrated stocks were prepared. Malachite green was usually added to the buffer stocks.
Sample PreparationDifferent concentrations were tested for malachite green and DNA templates. Components of a standard experiment are listed in the table below.
In each run up to 4 samples are measured simultaneously. Components that are the same in each of the 4 samples (buffer, DTT, NTPs, RPO, Water) are prepared as a 4.1x MasterMix in a loBind tube and split to the 4 cuvettes. Final Volume of each sample is 100 µl. Cary EclipseFor fluorescence measurements, a Cary Eclipse Spectrofluorimeter with a Multicellholder (4 cells) is used. Kinetics are recorded at 37° C. Excitation wavelength is 630 nm, emission is followed at 650 nm and 655 nm. After each kinetics measurement, spectra are to be recorded. CloseReferences[1] http://www.promega.com/catalog/catalogproducts.aspx?categoryname=productleaf_335&ckt=1 [2] Zubay, G. (1980) Meth. Enzymol. 65, 856–77, Zubay, G. (1973) Ann. Rev. Genet. 7, 267–87. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"