Team:TU Munich/Software
From 2010.igem.org
(→Tutorial) |
|||
| (115 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
<!-- ############## WIKI-PAGE STARTS HERE ############## --> | <!-- ############## WIKI-PAGE STARTS HERE ############## --> | ||
| - | |||
=Motivation= | =Motivation= | ||
| - | + | As described in the [[Team:TU_Munich/Project | project]] section, our vision is to provide a platform that allows to '''connect BioBricks'''. | |
| - | + | <br> <br> | |
| + | Although we have not yet been able to create a reliable and robust switch in the lab, we still want to demonstrate the power and simplicity of our approach: The following program (written in Java) allows the user to put together a network of logic gates and BioBricks as input and output. Here, we give the user the choice between a few input and ouput BioBricks. In principle, any (of at least 200) transcription-regulating BioBrick can be used as an input, and any gene can serve as an output. After entering the network, the software uses our current designs for AND, OR and NOT gates to generate one single DNA plasmid that contains the custom network - including all logical elements. | ||
| + | <br> <br> | ||
| + | '''Our vision''': Put in your logical network and get out your DNA implementation! | ||
| - | =Applet= | + | =Program= |
| + | <html><p>Click here to <a href="https://2010.igem.org/Team:TU_Munich/Software/Applet" target="java_applet">start the program </a> (loading may take a few seconds during which an empty wiki page is displayed).</p></html> | ||
| + | <br> | ||
| + | If you use the software for the first time, you might want to take a look at the [[Team:TU_Munich/Software#Tutorial | tutorial]] or the [[Team:TU_Munich/Software#Examples | step-by-step examples]] on how to use the program.<br> | ||
| + | <br> | ||
| + | Feel free to [[Media:TUM2010_Software_src.zip | download the source code]] of our open source program. | ||
| + | <br><br> | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart1}}More information about the Biobricks used within the program{{:Team:TU Munich/Templates/ToggleBoxStart2}}{{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
| + | The plasmid generated by the program is based on the [http://partsregistry.org/Part:pSB1A3 pSB1A3 backbone] which is a frequently-used plasmid carrying an ampicillin resistance. The design used for the logic gates is based on the His-Terminator design (see [[Team:TU_Munich/Project | project]] for details). If required for output proteins the ribosome binding site [http://partsregistry.org/Part:BBa_B0034 B0034] was used. For the constitutive promoter, positioned upstream of every logic gate, the software uses the [http://partsregistry.org/Part:BBa_J23100 J23100] promoter. Details about input and output Biobricks are supplied within the program. For all input BioBricks, the program only incorporates the target DNA sequence of the corresponding BioBrick. If a separate protein is required to bind the DNA, please note that this protein will not be included on the plasmid. | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | <br> | ||
| + | =Tutorial= | ||
| - | + | The program consists of three modules: '''Construct''', '''Verify''' and '''Synthesize'''. The first allows the user to enter inputs, whereas the second and third supply the user with information about the network. | |
| - | |||
| - | + | '''Construct''': Is the first module that enables the user to provide the inputs and outputs as well as the logic relations (AND, OR, NOT) to build a logic network to 'connect' inputs and outputs. The nods of the network correspond with the logic relations and the edges with the values 'true' or 'false'. | |
| - | + | ||
| + | '''Verify''': This module illustrates in the colors green for true and red for false which nods and relations are assigned the values 'true' or 'false'. | ||
| - | ''' | + | '''Sythesize''': This module assigns the abstract logic network concrete sequences, e.g. terminator sequences, generates random transmitter sequences and provides all the genetic information to rebuild the network. |
| - | + | ||
| - | + | ||
The different modules are described in detail below. | The different modules are described in detail below. | ||
| - | + | <br><br> | |
| - | + | *1. Step: Construct a logic network | |
| - | + | ||
{{:Team:TU Munich/Templates/ToggleBoxStart}} | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| - | A '''logic network''' can be designed by adding | + | A '''logic network''' can be designed by adding gates (via double clicking) and interconnecting them (via selecting two gates and pressing the 'C' key). |
| - | There are five different types of | + | There are five different types of gates: '''inputs''', '''outputs''', '''AND'''-gates, '''OR'''-gates and '''NOT'''-gates. The type of a gate can be changed by clicking on the combo box on the gate and selecting a type. Gates can be interconnected by selecting (via clicking inside the area of a gate) two gates and then pressing the C key (for connect). A complete logic network has to have at least one input and at least one output. Different gates may require different numbers of ingoing and outgoing connections (e.g. a NOT-gate requires one ingoing connection and allows for an arbitrary number of outgoing connections, whereas the AND-gate requires two ingoing connections and allows for a arbitrary number of outgoing connections; an input may not have an incoming connection, an output no outgoing connection). In addition the logic network may not include loops. |
| - | The user has to assign DNA sequences (either custom RNA sequences or sequences coding for proteins) to all input and output | + | The user has to assign DNA sequences (promoter sequences or either custom RNA sequences or sequences coding for proteins) to all input and output gates by clicking on the "Set input" button of the gates. |
| - | Invalid inputs (e.g. a letter different from G, C, T, A or N in a nucleotide sequence; loop(s) in the logic network; no sequences assigned to input(s)/output(s); input or output sequences that are too similar) are reported back to the users as error messages providing a description of the errors and advices to correct the errors. The Check and Turn-to-DNA modules can only be entered when the network contains '''no errors'''. | + | Invalid inputs (e.g. a letter different from G, C, T, A or N in a nucleotide sequence; loop(s) in the logic network; no sequences assigned to input(s)/output(s); input or output sequences that are too similar) are reported back to the users as error messages providing a description of the errors and advices how to correct the errors. The Check and Turn-to-DNA modules can only be entered when the network contains '''no errors'''. |
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| - | + | *2. Step: Verify the logic network | |
| - | + | ||
{{:Team:TU Munich/Templates/ToggleBoxStart}} | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
The logic network designed in step 1 can now be verified. | The logic network designed in step 1 can now be verified. | ||
| - | Logical values are represented by colors (green = true, red = false). Thus a green box represents a | + | Logical values are represented by colors (green = true, red = false). Thus a green box represents a gates (e.g. an AND-gate) with the logical value true, a red box a gates with the logical value false. The flow of logical value can be followed by following the colored edges between the gates. |
The logical value(s) of the input(s) can be manipulated by clicking on their checkbox(es). | The logical value(s) of the input(s) can be manipulated by clicking on their checkbox(es). | ||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| - | + | *3. Step: Synthesize - Turn the logic network into DNA | |
| - | + | ||
{{:Team:TU Munich/Templates/ToggleBoxStart}} | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
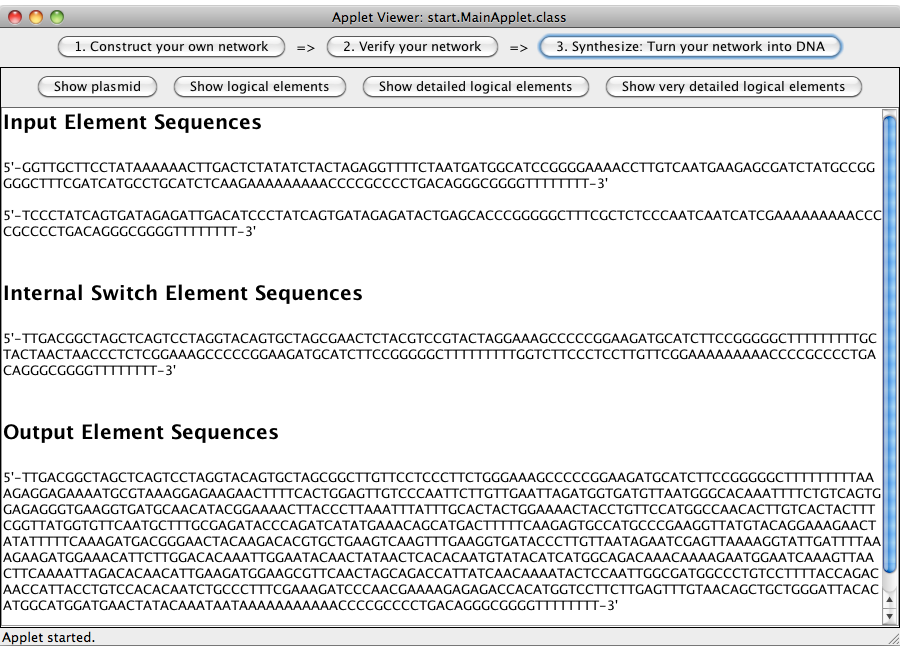
| - | A set of genetic | + | A set of genetic gates (categorized as input, logic gates and output) representing the logical network designed in step 1 is calculated and displayed. |
| + | |||
| + | All elements consist of a promoter, a ribosome binding site, up to two toggle switches, one output transmitter sequences and at least one terminator. | ||
| + | <br><br> | ||
| + | This information may at a later point, when the functionality of our toggle switches has been demonstrated in the lab and not only in silicio, be used for synthesizing the genetic networks. | ||
| - | |||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | =Examples= | ||
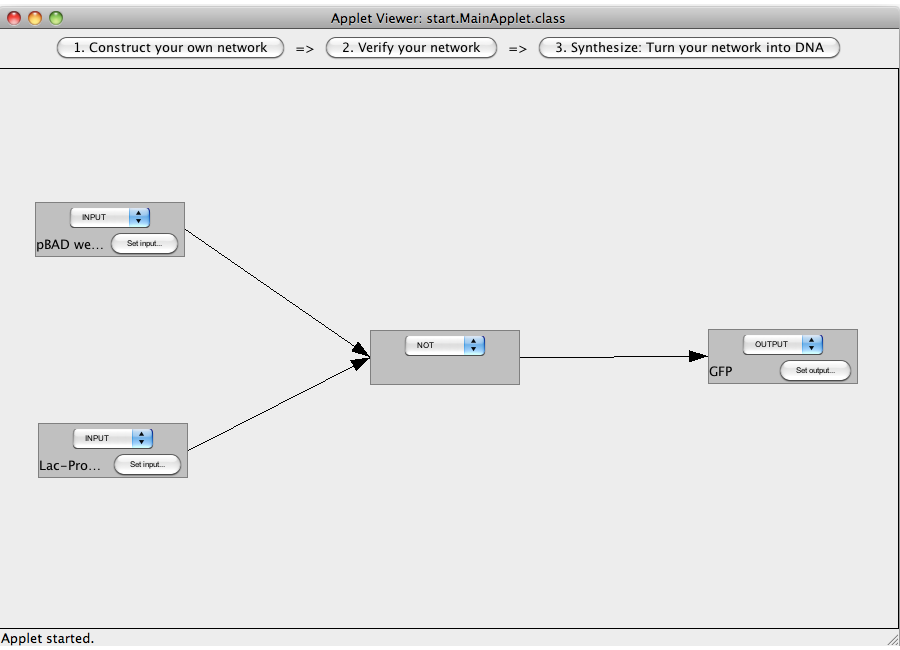
| - | == | + | ==A NOT-gate (introductory)== |
| + | The following "Read more" section will provide step-by-step instructions for creating a simple network, connecting two Biobricks with a NOT gate. | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | The following "Read more" section will provide step-by-step instructions for creating a simple network, connecting two Biobricks with a NOT gate. | ||
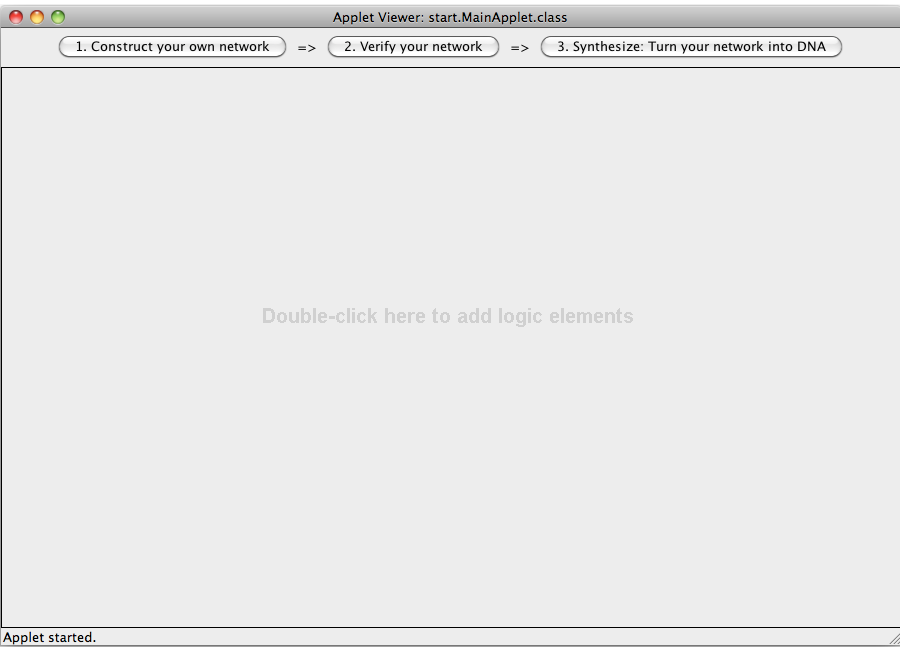
| + | [[Image:Software_NOT_01.png|600px|thumb|center|Step 01: After starting the program this screen is displayed. It has of three views: Construct, Verify and Synthesize. The later two can only be entered if a valid logic network has been constructed.]]<br> | ||
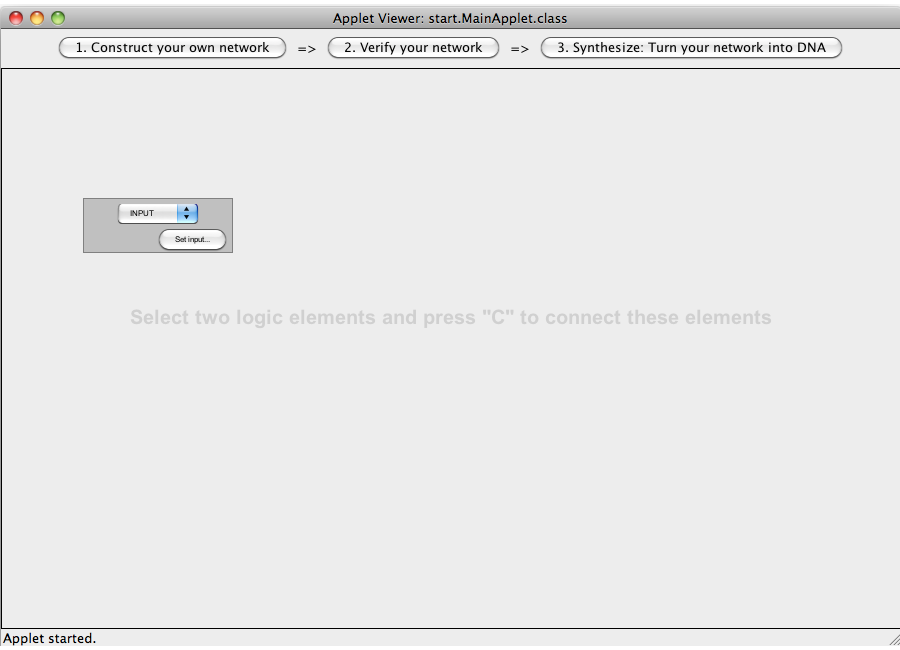
| + | [[Image:Software_NOT_02.png|600px|thumb|center|Step 02: Add a new gate via double clicking.]]<br> | ||
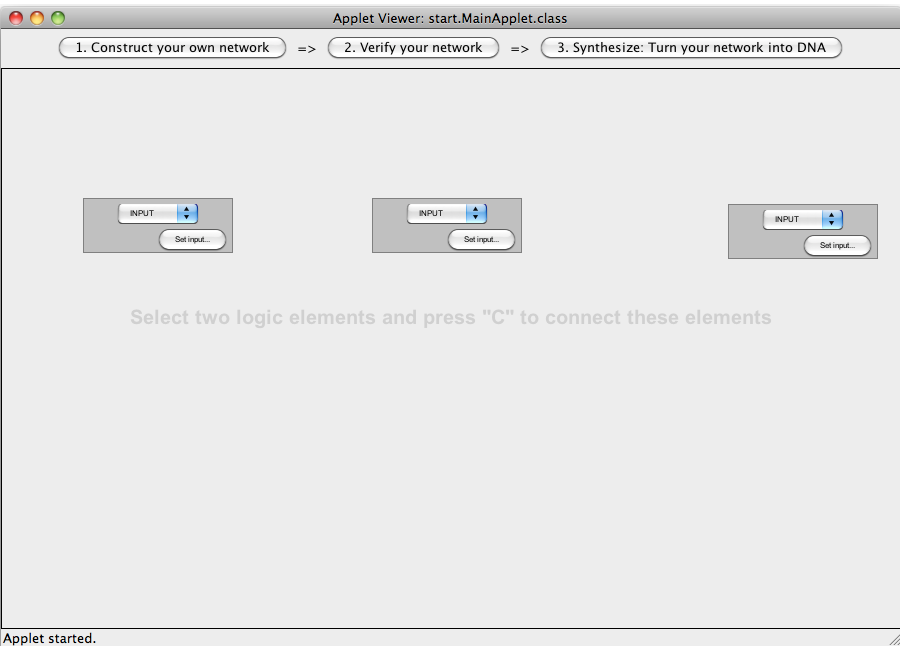
| + | [[Image:Software_NOT_03.png|600px|thumb|center|Step 03: Add two more gates in the same way.]]<br> | ||
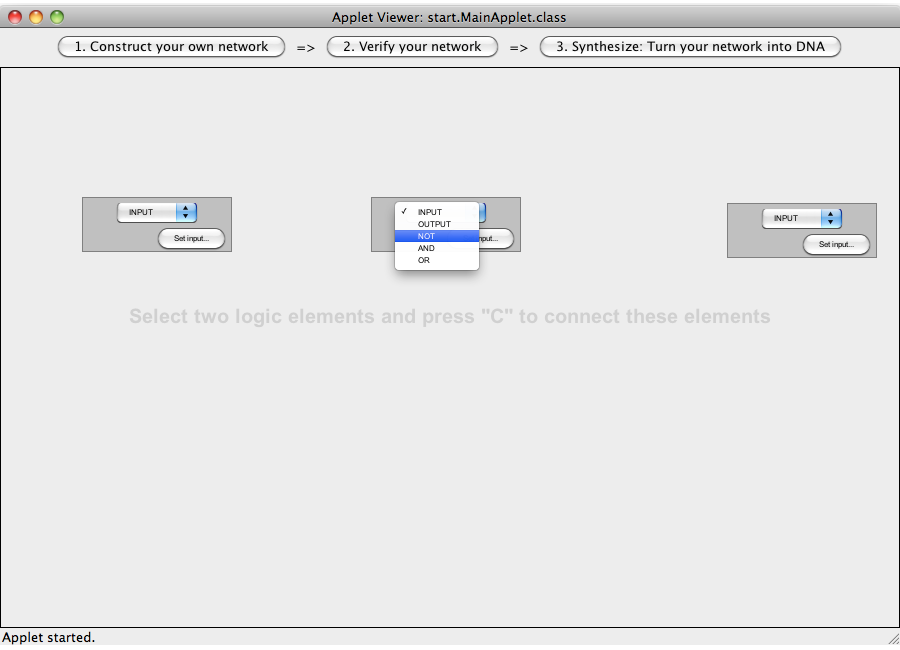
| + | [[Image:Software_NOT_04.png|600px|thumb|center|Step 04: You can change the type of the logic gates via the combo box]]<br> | ||
| + | [[Image:Software_NOT_05.png|600px|thumb|center|Step 05: Change the type to: Input, Not, Output.]]<br> | ||
| + | [[Image:Software_NOT_06.png|600px|thumb|center|Step 06: Clicking inside the area of a gate selects it (indicated by the blue color). You can select as many logical gates as you like. To deselect, click on the empty space between the gates. Once a gate is selected you can delete it by pressing the DEL or BACKSPACE key.]]<br> | ||
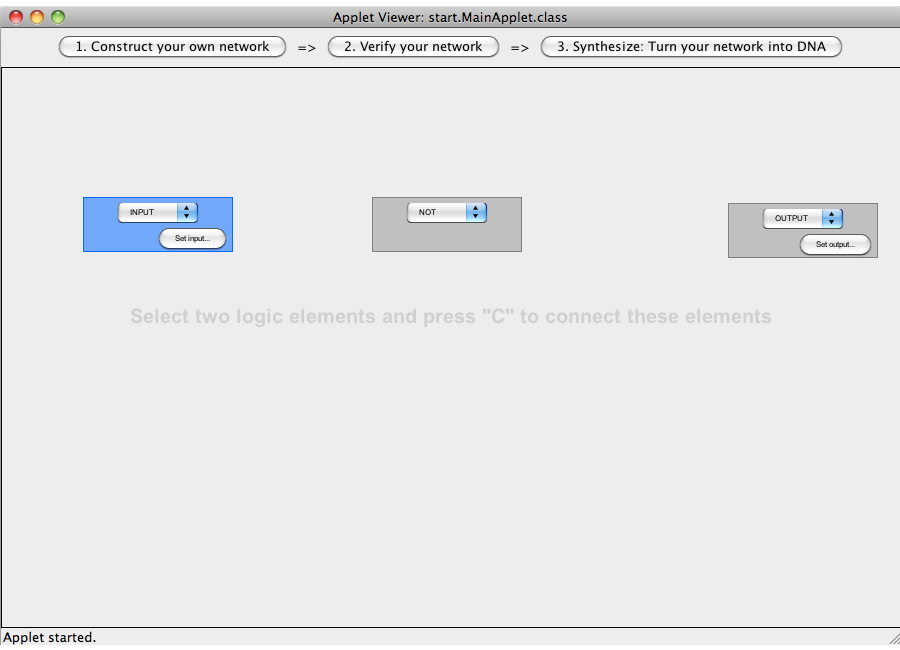
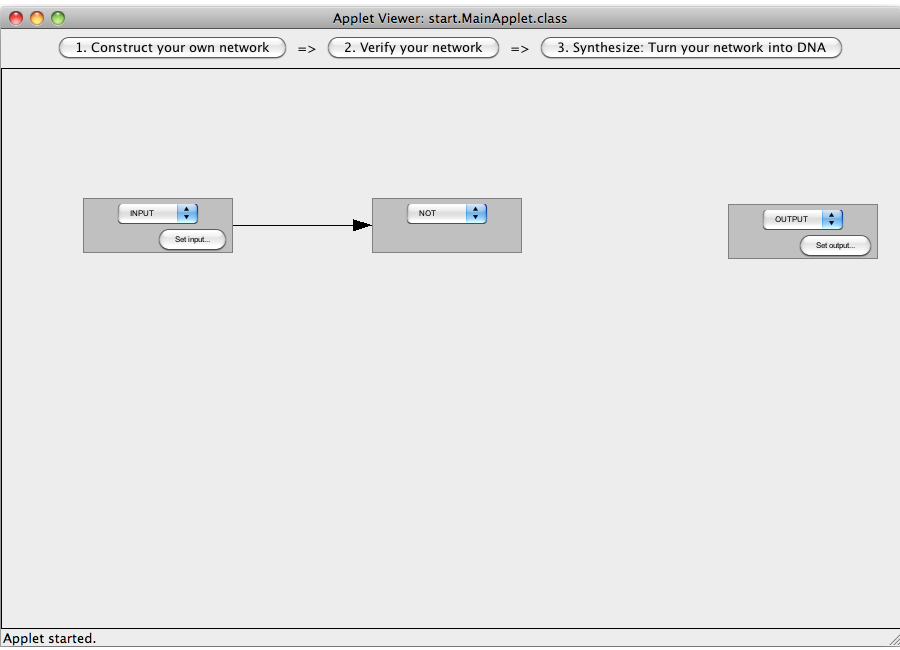
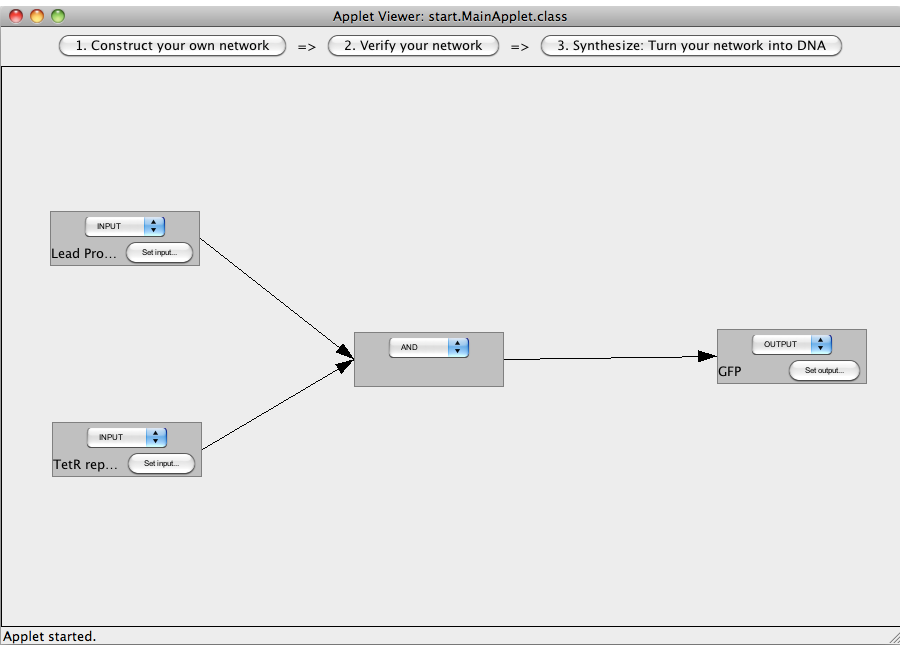
| + | [[Image:Software_NOT_07.png|600px|thumb|center|Step 07: To connect two gates select both. Note that the connections has a direction (which will be indicated by an arrow), thus you have to select the gate in the correct order: first, select the box from which the connection starts and then select the box where the connection ends (The arrow points from the box that generates an output transmitter molecule, to the target box that will be activated by this output transmitter). You can only connect gates pairwise.]]<br> | ||
| + | [[Image:Software_NOT_08.png|600px|thumb|center|Step 08: Two gates have been connected.]]<br> | ||
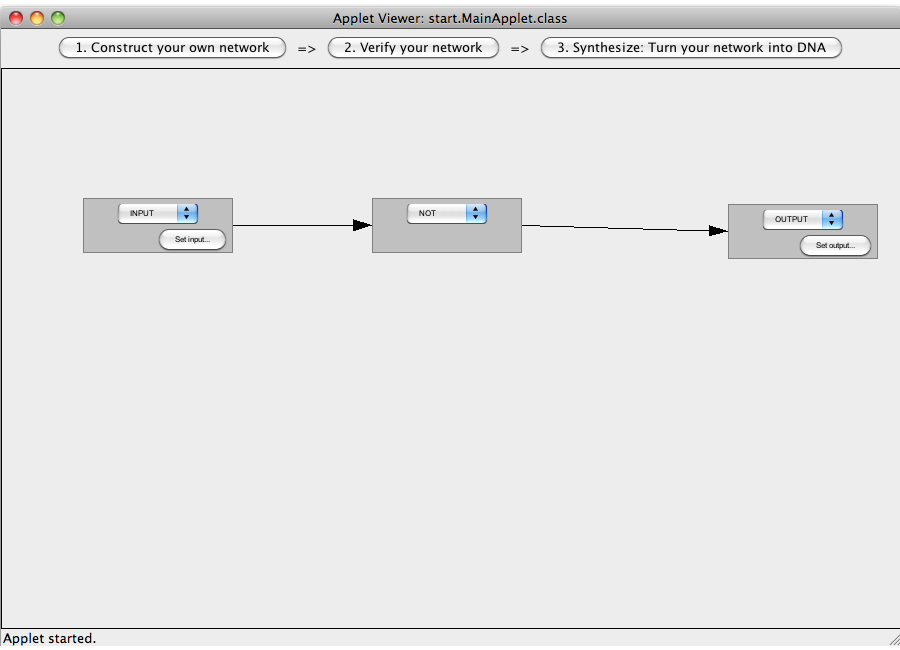
| + | [[Image:Software_NOT_09.png|600px|thumb|center|Step 09: Connected the whole network.]]<br> | ||
| + | [[Image:Software_NOT_10.png|600px|thumb|center|Step 10: Press the "Set input ..." button on the input to select a promoter that controls the input.]]<br> | ||
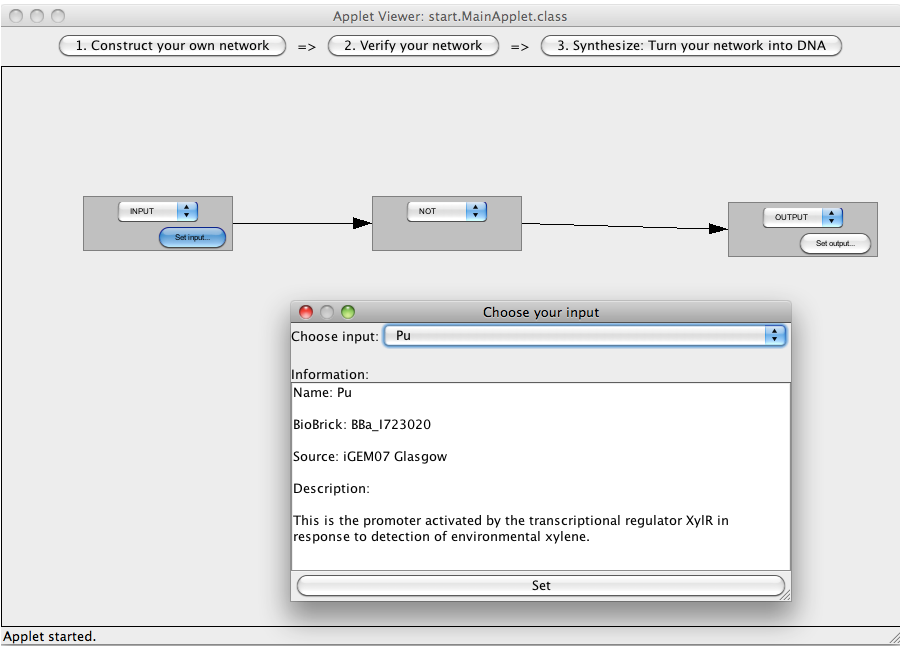
| + | [[Image:Software_NOT_11.png|600px|thumb|center|Step 11: Choose one of the exemplary chosen BioBricks and press set.]]<br> | ||
| + | [[Image:Software_NOT_12.png|600px|thumb|center|Step 12: Press the "Set output ..." button on the output to select an output.]]<br> | ||
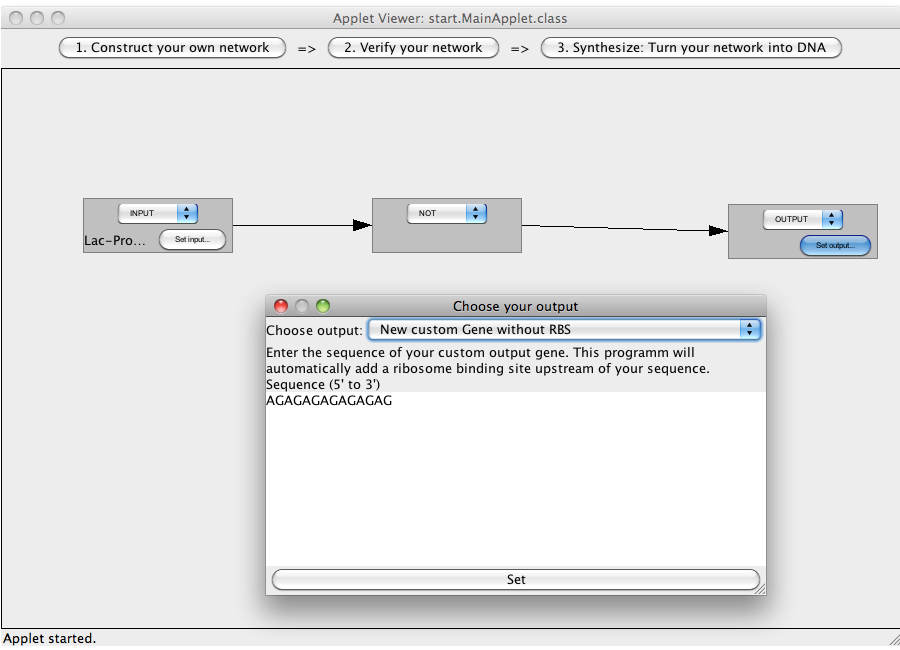
| + | [[Image:Software_NOT_13.png|600px|thumb|center|Step 13: You can chose a BioBrick or enter a custom made sequence (DNA with or without RBS, RNA).]]<br> | ||
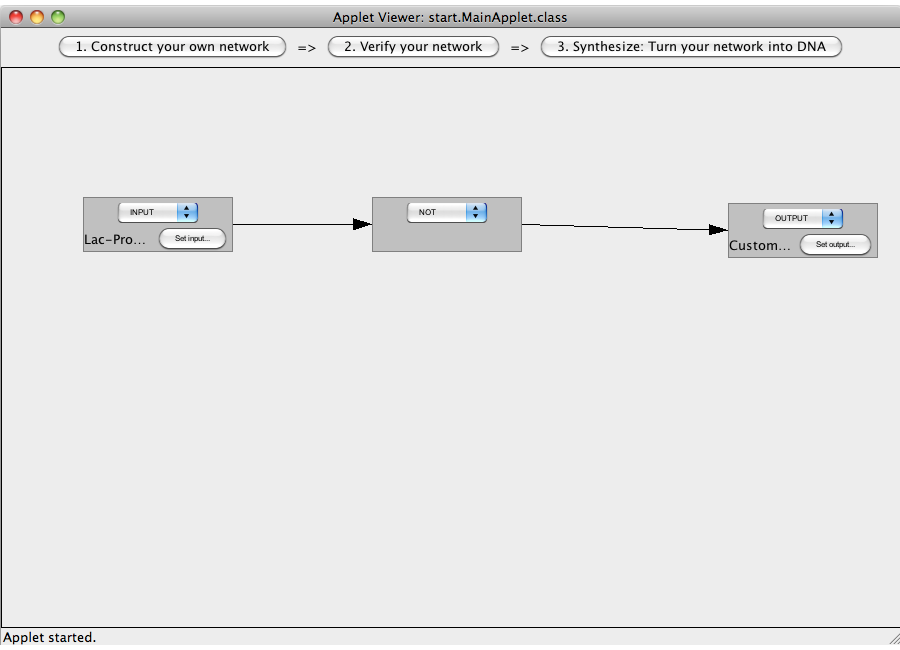
| + | [[Image:Software_NOT_14.png|600px|thumb|center|Step 14: The logic network has been constructed. Press on the Button "2. Verify your network" to leave the construction view. If you have made mistakes during the construction process, an error message will be displayed.]]<br> | ||
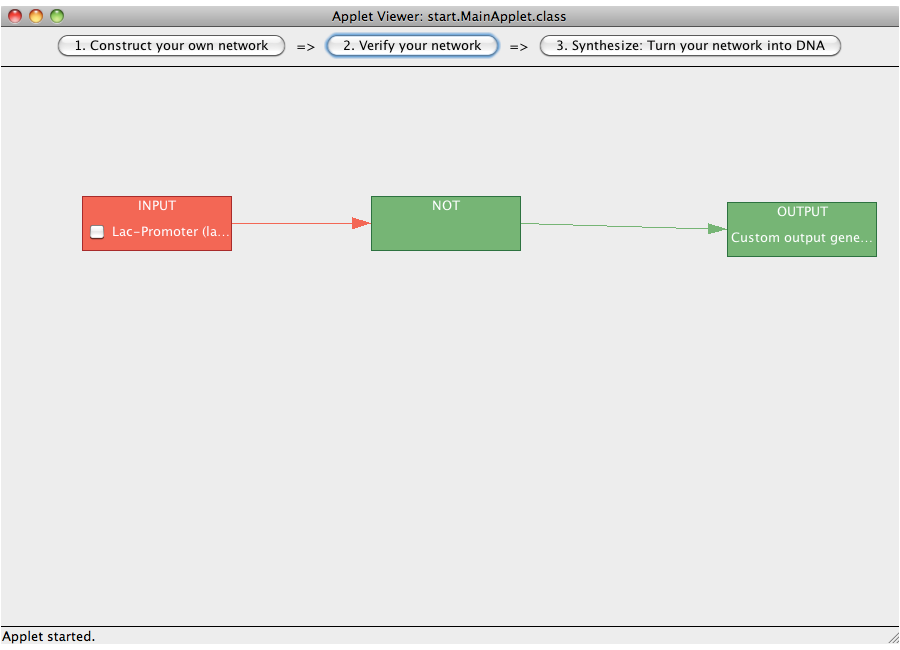
| + | [[Image:Software_NOT_15.png|600px|thumb|center|Step 15: Check your network.]]<br> | ||
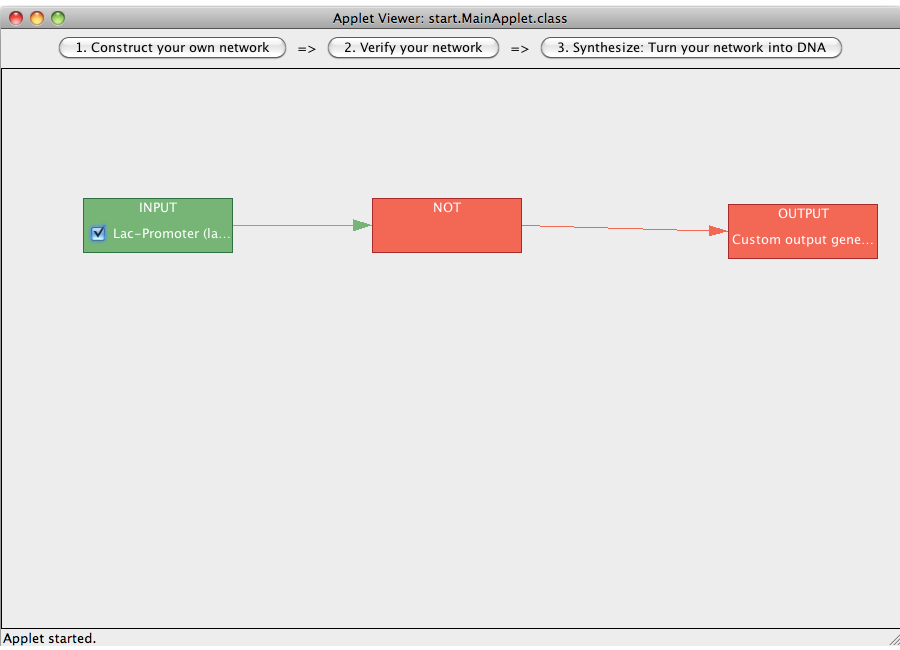
| + | [[Image:Software_NOT_16.png|600px|thumb|center|Step 16: By selecting or deselecting the checkbox on the inputs one can study the flow of logical value through the network.]]<br> | ||
| + | [[Image:Software_NOT_17.png|600px|thumb|center|Step 17: Press "Synthesize" and the logic network gets turned into DNA. The resulting DNA can get represented in four different views depending on the level of detail you wish to see.]]<br> | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
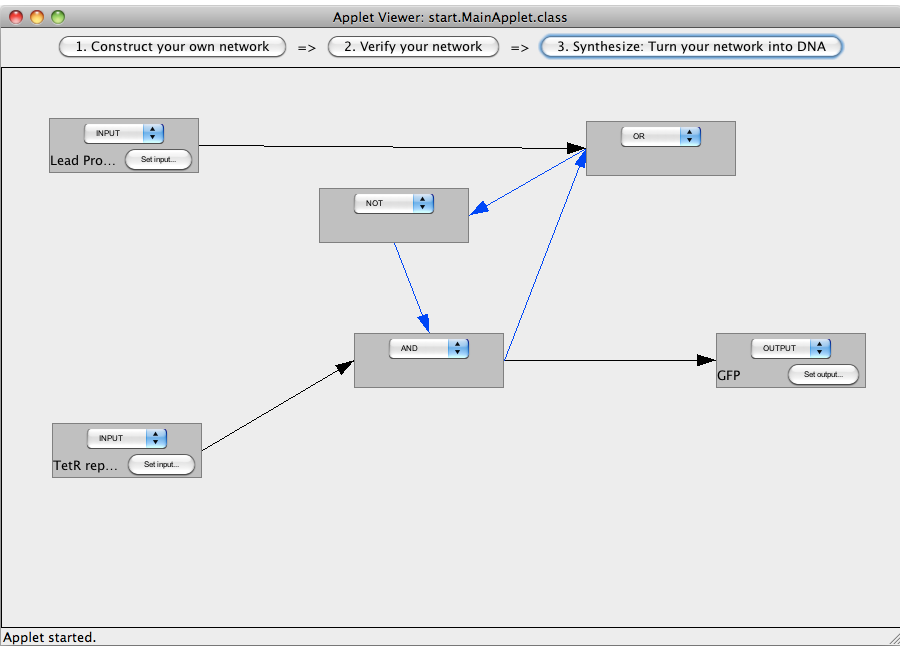
| + | ==An AND-gate== | ||
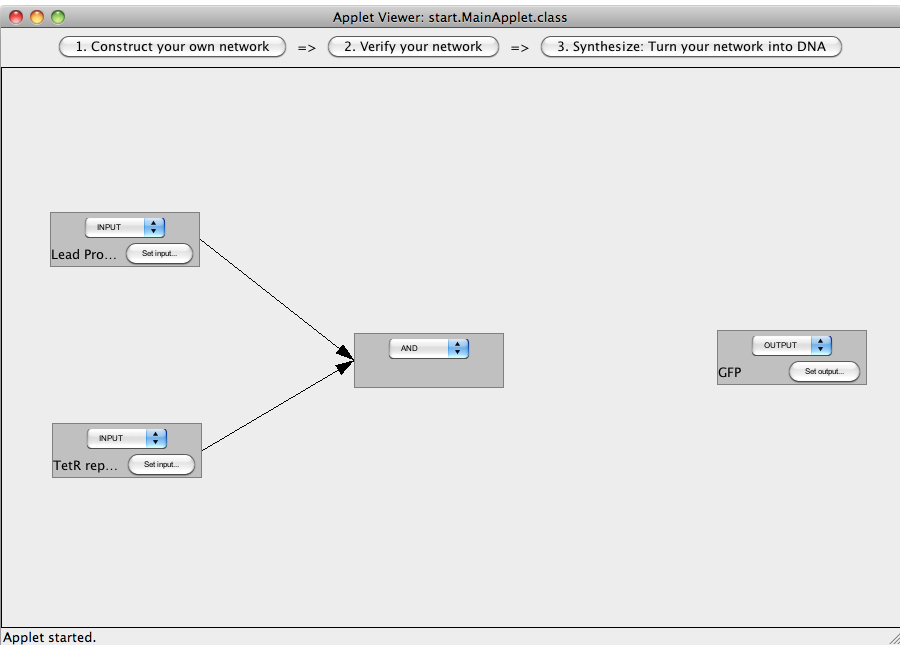
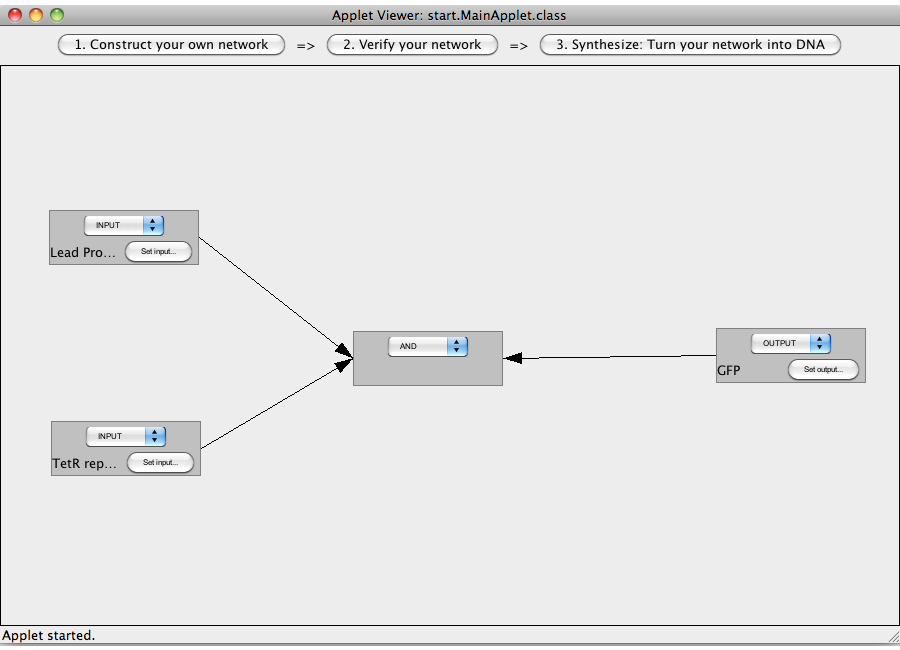
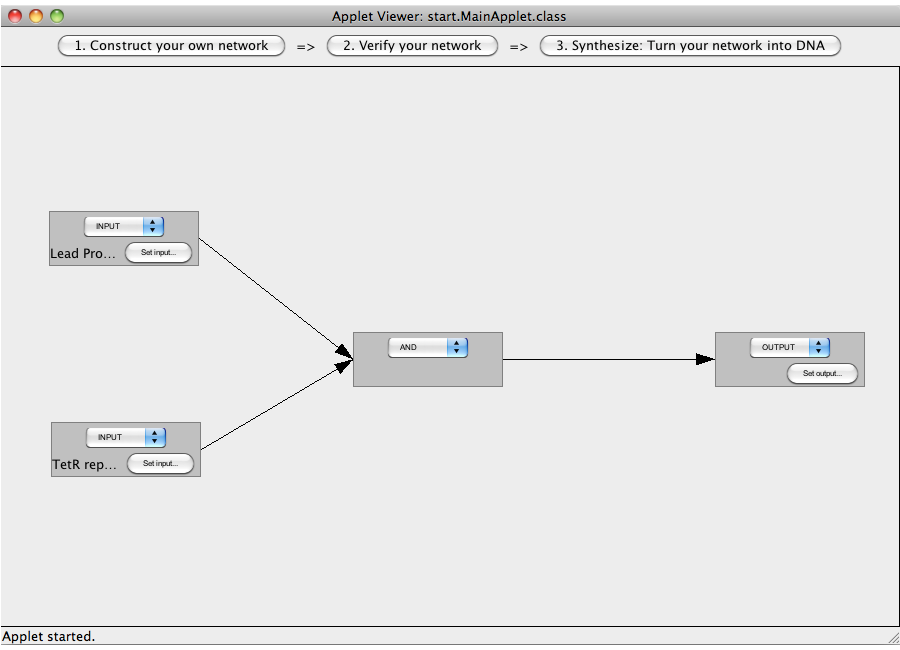
| + | This tutorial will use an AND gate connecting three Biobricks. | ||
{{:Team:TU Munich/Templates/ToggleBoxStart}} | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| - | + | [[Image:Software_AND_01.png|600px|thumb|center|Step 01: Add four gates and set their types.]]<br> | |
| + | [[Image:Software_AND_02.png|600px|thumb|center|Step 02: Set the input and output sequences.]]<br> | ||
| + | [[Image:Software_AND_03.png|600px|thumb|center|Step 03: Interconnect the gates.]]<br> | ||
| + | [[Image:Software_AND_04.png|600px|thumb|center|Step 04: Check the flow of logic value.]]<br> | ||
| + | [[Image:Software_AND_05.png|600px|thumb|center|Step 05: Synthesize the network.]]<br> | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | |||
| + | ==Common mistakes== | ||
| + | In case you got stuck with a problem, you might find the solution among the following list of common mistakes. | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | [[Image:Software_CommonMistakes_01.png|600px|thumb|center|The logic network is not connected to an output.]]<br> | ||
| + | [[Image:Software_CommonMistakes_02.png|600px|thumb|center|A connection has been established in the wrong direction.]]<br> | ||
| + | [[Image:Software_CommonMistakes_03.png|600px|thumb|center|A input / output sequence is not set.]]<br> | ||
| + | [[Image:Software_CommonMistakes_04.png|600px|thumb|center|The network contains a loop.]]<br> | ||
| + | [[Image:Software_CommonMistakes_05.png|600px|thumb|center|A gate has too many / not enough connections.]]<br> | ||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| Line 78: | Line 126: | ||
(INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS | (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS | ||
SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE. | SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE. | ||
| - | + | <br>(Inspired by: [http://en.wikipedia.org/wiki/BSD_licenses BSD license]) | |
| - | + | ||
| - | (Inspired by: [http://en.wikipedia.org/wiki/BSD_licenses | + | |
==Biology== | ==Biology== | ||
THIS SOFTWARE IS FOR '''DEMONSTRATION PURPOSES ONLY'''. '''THE WARRANTY''' OF FITNESS FOR A PARTICULAR PURPOSE, INCLUDING THE DESIGN OF FUNCTIONAL GENE CIRCUITS, '''IS DISCLAIMED'''. ESPECIALLY '''DO NOT''' USE THE SOFTWARE IN THE PROCESS OF '''DESIGNING REAL EXPERIMENTS''' OR '''ORDERING GENETIC DEVICES / SEQUENCES'''. | THIS SOFTWARE IS FOR '''DEMONSTRATION PURPOSES ONLY'''. '''THE WARRANTY''' OF FITNESS FOR A PARTICULAR PURPOSE, INCLUDING THE DESIGN OF FUNCTIONAL GENE CIRCUITS, '''IS DISCLAIMED'''. ESPECIALLY '''DO NOT''' USE THE SOFTWARE IN THE PROCESS OF '''DESIGNING REAL EXPERIMENTS''' OR '''ORDERING GENETIC DEVICES / SEQUENCES'''. | ||
| - | |||
| - | |||
| - | |||
| - | |||
Latest revision as of 17:39, 20 November 2010
|
||||||||
|
|
MotivationAs described in the project section, our vision is to provide a platform that allows to connect BioBricks.
ProgramClick here to start the program (loading may take a few seconds during which an empty wiki page is displayed). If you use the software for the first time, you might want to take a look at the tutorial or the step-by-step examples on how to use the program. Feel free to download the source code of our open source program. More information about the Biobricks used within the program The plasmid generated by the program is based on the [http://partsregistry.org/Part:pSB1A3 pSB1A3 backbone] which is a frequently-used plasmid carrying an ampicillin resistance. The design used for the logic gates is based on the His-Terminator design (see project for details). If required for output proteins the ribosome binding site [http://partsregistry.org/Part:BBa_B0034 B0034] was used. For the constitutive promoter, positioned upstream of every logic gate, the software uses the [http://partsregistry.org/Part:BBa_J23100 J23100] promoter. Details about input and output Biobricks are supplied within the program. For all input BioBricks, the program only incorporates the target DNA sequence of the corresponding BioBrick. If a separate protein is required to bind the DNA, please note that this protein will not be included on the plasmid. CloseTutorialThe program consists of three modules: Construct, Verify and Synthesize. The first allows the user to enter inputs, whereas the second and third supply the user with information about the network.
Verify: This module illustrates in the colors green for true and red for false which nods and relations are assigned the values 'true' or 'false'. Sythesize: This module assigns the abstract logic network concrete sequences, e.g. terminator sequences, generates random transmitter sequences and provides all the genetic information to rebuild the network. The different modules are described in detail below.
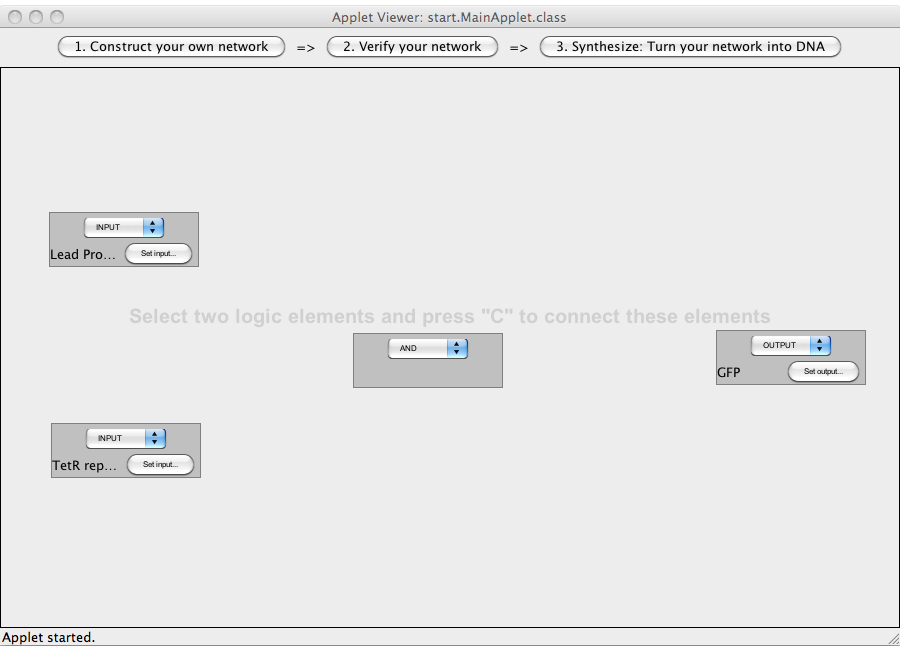
A logic network can be designed by adding gates (via double clicking) and interconnecting them (via selecting two gates and pressing the 'C' key). There are five different types of gates: inputs, outputs, AND-gates, OR-gates and NOT-gates. The type of a gate can be changed by clicking on the combo box on the gate and selecting a type. Gates can be interconnected by selecting (via clicking inside the area of a gate) two gates and then pressing the C key (for connect). A complete logic network has to have at least one input and at least one output. Different gates may require different numbers of ingoing and outgoing connections (e.g. a NOT-gate requires one ingoing connection and allows for an arbitrary number of outgoing connections, whereas the AND-gate requires two ingoing connections and allows for a arbitrary number of outgoing connections; an input may not have an incoming connection, an output no outgoing connection). In addition the logic network may not include loops. The user has to assign DNA sequences (promoter sequences or either custom RNA sequences or sequences coding for proteins) to all input and output gates by clicking on the "Set input" button of the gates.
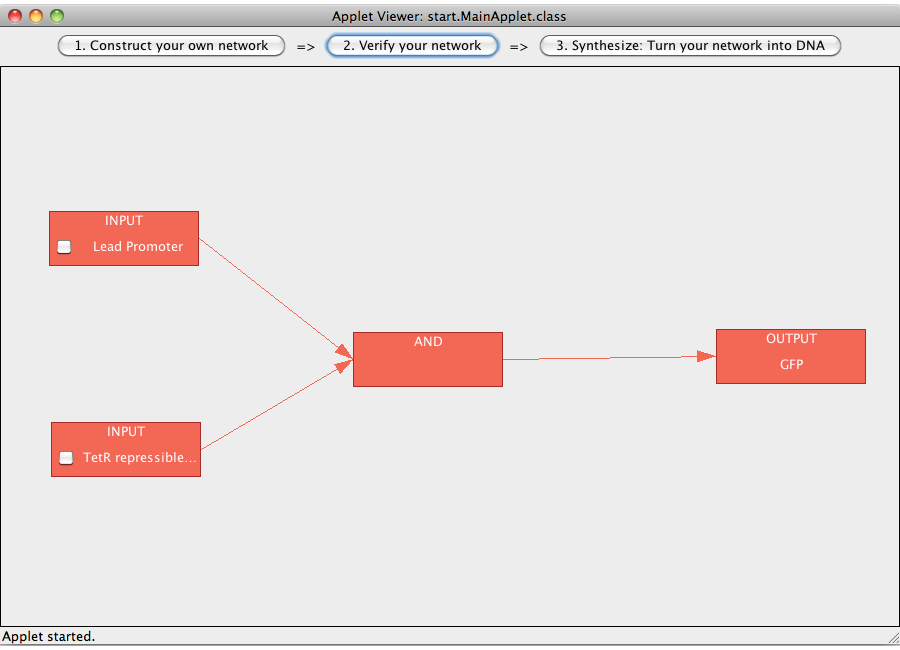
The logic network designed in step 1 can now be verified. Logical values are represented by colors (green = true, red = false). Thus a green box represents a gates (e.g. an AND-gate) with the logical value true, a red box a gates with the logical value false. The flow of logical value can be followed by following the colored edges between the gates. The logical value(s) of the input(s) can be manipulated by clicking on their checkbox(es). Close
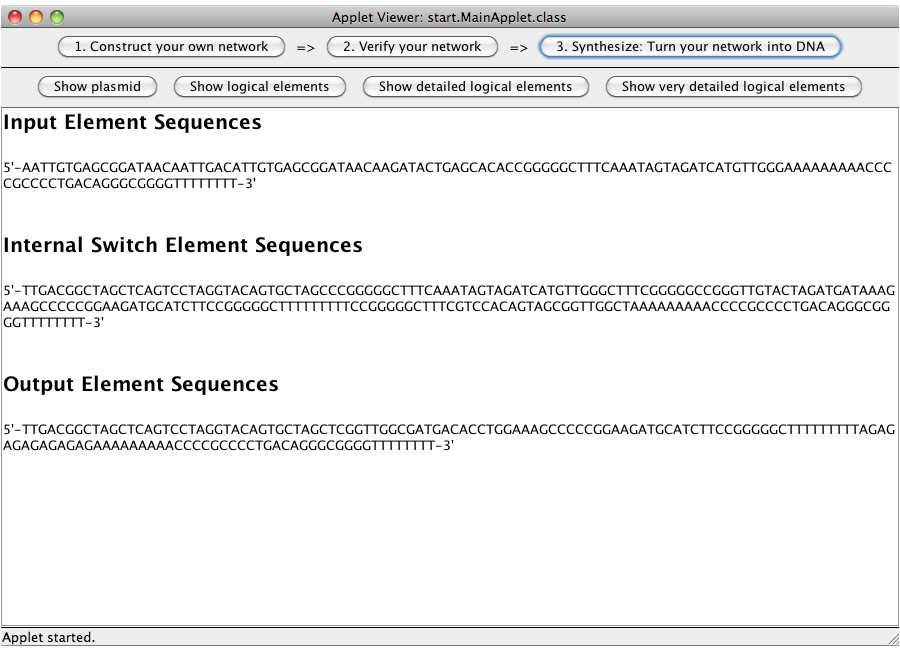
A set of genetic gates (categorized as input, logic gates and output) representing the logical network designed in step 1 is calculated and displayed. All elements consist of a promoter, a ribosome binding site, up to two toggle switches, one output transmitter sequences and at least one terminator.
ExamplesA NOT-gate (introductory)The following "Read more" section will provide step-by-step instructions for creating a simple network, connecting two Biobricks with a NOT gate. The following "Read more" section will provide step-by-step instructions for creating a simple network, connecting two Biobricks with a NOT gate. 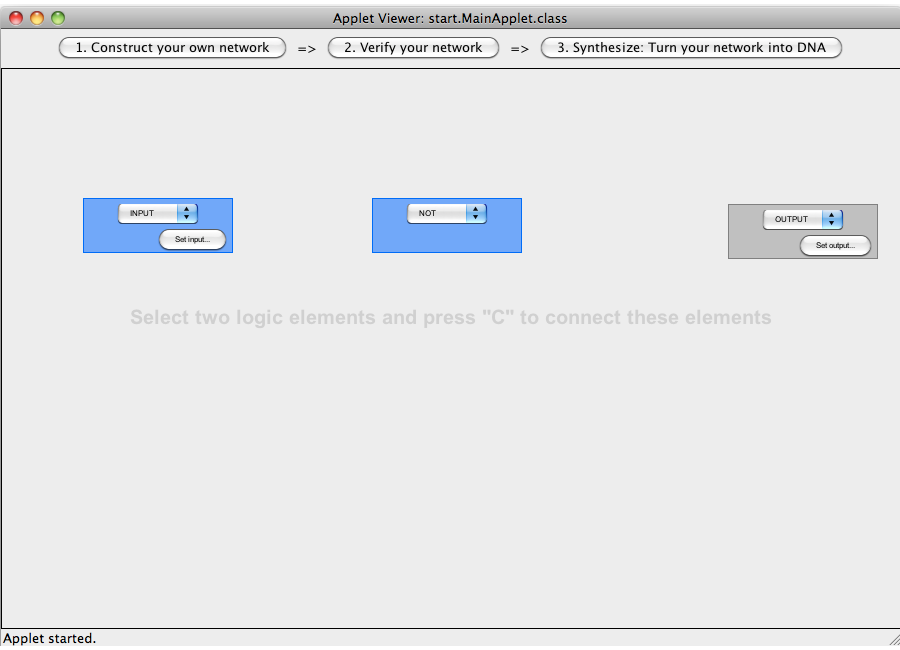 Step 07: To connect two gates select both. Note that the connections has a direction (which will be indicated by an arrow), thus you have to select the gate in the correct order: first, select the box from which the connection starts and then select the box where the connection ends (The arrow points from the box that generates an output transmitter molecule, to the target box that will be activated by this output transmitter). You can only connect gates pairwise. Close An AND-gateThis tutorial will use an AND gate connecting three Biobricks. Common mistakesIn case you got stuck with a problem, you might find the solution among the following list of common mistakes. DisclaimerSoftwareTHIS SOFTWARE IS PROVIDED BY THE CONTRIBUTORS "AS IS" AND
ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED
WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
DISCLAIMED. IN NO EVENT SHALL THE CONTRIBUTORS BE LIABLE FOR ANY
DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES
(INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES;
LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND
ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT
(INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS
SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
BiologyTHIS SOFTWARE IS FOR DEMONSTRATION PURPOSES ONLY. THE WARRANTY OF FITNESS FOR A PARTICULAR PURPOSE, INCLUDING THE DESIGN OF FUNCTIONAL GENE CIRCUITS, IS DISCLAIMED. ESPECIALLY DO NOT USE THE SOFTWARE IN THE PROCESS OF DESIGNING REAL EXPERIMENTS OR ORDERING GENETIC DEVICES / SEQUENCES.
|
|||||||
 "
"