Team:Imperial College London/Modelling
From 2010.igem.org
m |
|||
| (15 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
|- | |- | ||
| - | | | + | |During the design of our construct three major questions arose which could be answered by computer modelling: |
| + | |||
<ol> | <ol> | ||
| Line 13: | Line 14: | ||
|style="font-family: helvetica, arial, sans-serif;font-size:1.2em;color:#ea8828;" align="right"|[[Team:Imperial_College_London/Modelling/Protein_Display/Objectives|Objectives]] | [[Team:Imperial_College_London/Modelling/Protein_Display/Detailed_Description|Description]] | [[Team:Imperial_College_London/Modelling/Protein_Display/Parameters_and_Constants|Constants]] | [[Team:Imperial_College_London/Modelling/Protein_Display/Results_and_Conclusion|Results]] | [[Team:Imperial_College_London/Modelling/Protein_Display/Download_MatLab_Files|MATLAB Code]] | |style="font-family: helvetica, arial, sans-serif;font-size:1.2em;color:#ea8828;" align="right"|[[Team:Imperial_College_London/Modelling/Protein_Display/Objectives|Objectives]] | [[Team:Imperial_College_London/Modelling/Protein_Display/Detailed_Description|Description]] | [[Team:Imperial_College_London/Modelling/Protein_Display/Parameters_and_Constants|Constants]] | [[Team:Imperial_College_London/Modelling/Protein_Display/Results_and_Conclusion|Results]] | [[Team:Imperial_College_London/Modelling/Protein_Display/Download_MatLab_Files|MATLAB Code]] | ||
|} | |} | ||
| - | We came up with a novel idea of detecting organisms that we do not have a specific receptor for. In our particular example, the protease of Schistosoma is meant to cleave a protein displayed on the bacteria's cell wall. The cleaved peptide is | + | We came up with a novel idea of detecting organisms that we do not have a specific receptor for. In our particular example, the protease of Schistosoma is meant to cleave a protein displayed on the bacteria's cell wall (<i>the pink rectangle on the diagram below</i>). The cleaved peptide is recognized by the receptor which would then activate the colour expression. This solution raised questions about the risk of false positive or whether there are any chances for ComD receptors to be activated in the diluted environment. Modelling of this module would enable us to answer these questions.<br/><br/> |
{| style="width:800px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;" | {| style="width:800px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;" | ||
| - | |2. <b> | + | |2. <b>Signaling Model</b> |
|style="font-family: helvetica, arial, sans-serif;font-size:1.2em;color:#ea8828;" align="right"|[[Team:Imperial_College_London/Modelling/Signalling/Objectives|Objectives]] | [[Team:Imperial_College_London/Modelling/Signalling/Detailed_Description|Description]] | [[Team:Imperial_College_London/Modelling/Signalling/Parameters_and_Constants|Constants]] | [[Team:Imperial_College_London/Modelling/Signalling/Results_and_Conclusion|Results]] | [[Team:Imperial_College_London/Modelling/Signalling/Download_MatLab_Files|MATLAB Code]] | |style="font-family: helvetica, arial, sans-serif;font-size:1.2em;color:#ea8828;" align="right"|[[Team:Imperial_College_London/Modelling/Signalling/Objectives|Objectives]] | [[Team:Imperial_College_London/Modelling/Signalling/Detailed_Description|Description]] | [[Team:Imperial_College_London/Modelling/Signalling/Parameters_and_Constants|Constants]] | [[Team:Imperial_College_London/Modelling/Signalling/Results_and_Conclusion|Results]] | [[Team:Imperial_College_London/Modelling/Signalling/Download_MatLab_Files|MATLAB Code]] | ||
|} | |} | ||
| - | We decided to use the ComCDE | + | We decided to use the ComCDE signaling pathway from ''S.pneumoniae'' and so questions arose on whether it would work appropriately in ''B.subtilis''. We modelled this system to make sure that the signalling pathway would be working as anticipated (<i>the green rectangle on the diagram below</i>).<br/><br/> |
{| style="width:800px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;" | {| style="width:800px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;" | ||
|3. <b>Fast Response Model</b> | |3. <b>Fast Response Model</b> | ||
|style="font-family: helvetica, arial, sans-serif;font-size:1.2em;color:#ea8828;" align="right"|[[Team:Imperial_College_London/Modelling/Output/Objectives|Objectives]] | [[Team:Imperial_College_London/Modelling/Output/Detailed_Description|Description]] | [[Team:Imperial_College_London/Modelling/Output/Parameters_and_Constants|Constants]] | [[Team:Imperial_College_London/Modelling/Output/Results_and_Conclusion|Results]] | [[Team:Imperial_College_London/Modelling/Output/Download_MatLab_Files|MATLAB Code]] | |style="font-family: helvetica, arial, sans-serif;font-size:1.2em;color:#ea8828;" align="right"|[[Team:Imperial_College_London/Modelling/Output/Objectives|Objectives]] | [[Team:Imperial_College_London/Modelling/Output/Detailed_Description|Description]] | [[Team:Imperial_College_London/Modelling/Output/Parameters_and_Constants|Constants]] | [[Team:Imperial_College_London/Modelling/Output/Results_and_Conclusion|Results]] | [[Team:Imperial_College_London/Modelling/Output/Download_MatLab_Files|MATLAB Code]] | ||
|} | |} | ||
| - | We came up with | + | We came up with the idea of using the amplification of a colour output, which would show within minutes of the stimulus being added (<i>the blue rectangle on the diagram below</i>). The question that arose was whether amplification will actually perform better than simple production (i.e. transcription and translation) in the cellular environment. Furthermore, we had difficulties deciding whether we should design the amplification module to consist of 1,2 or even more amplification steps. These issues were quite difficult to answer, so we decided to employ modelling.<br/> |
</ol> | </ol> | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:404px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:IC_Overview.jpg|400px]] | ||
| + | |- | ||
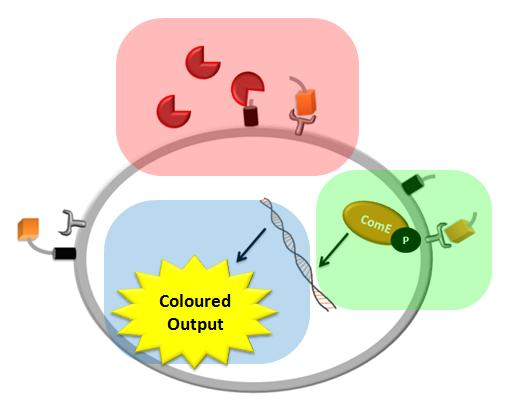
| + | |Each of the shadowed rectangles corresponds to the model of a particular module of the system. Pink corresponds to the model of the detection mechanism, green corresponds to the model of the signal transduction and blue corresponds to the model of the fast response module. | ||
| + | |} | ||
| + | </div> | ||
|} | |} | ||
| Line 33: | Line 42: | ||
|<b>Detection Model</b><br/> | |<b>Detection Model</b><br/> | ||
<ol> | <ol> | ||
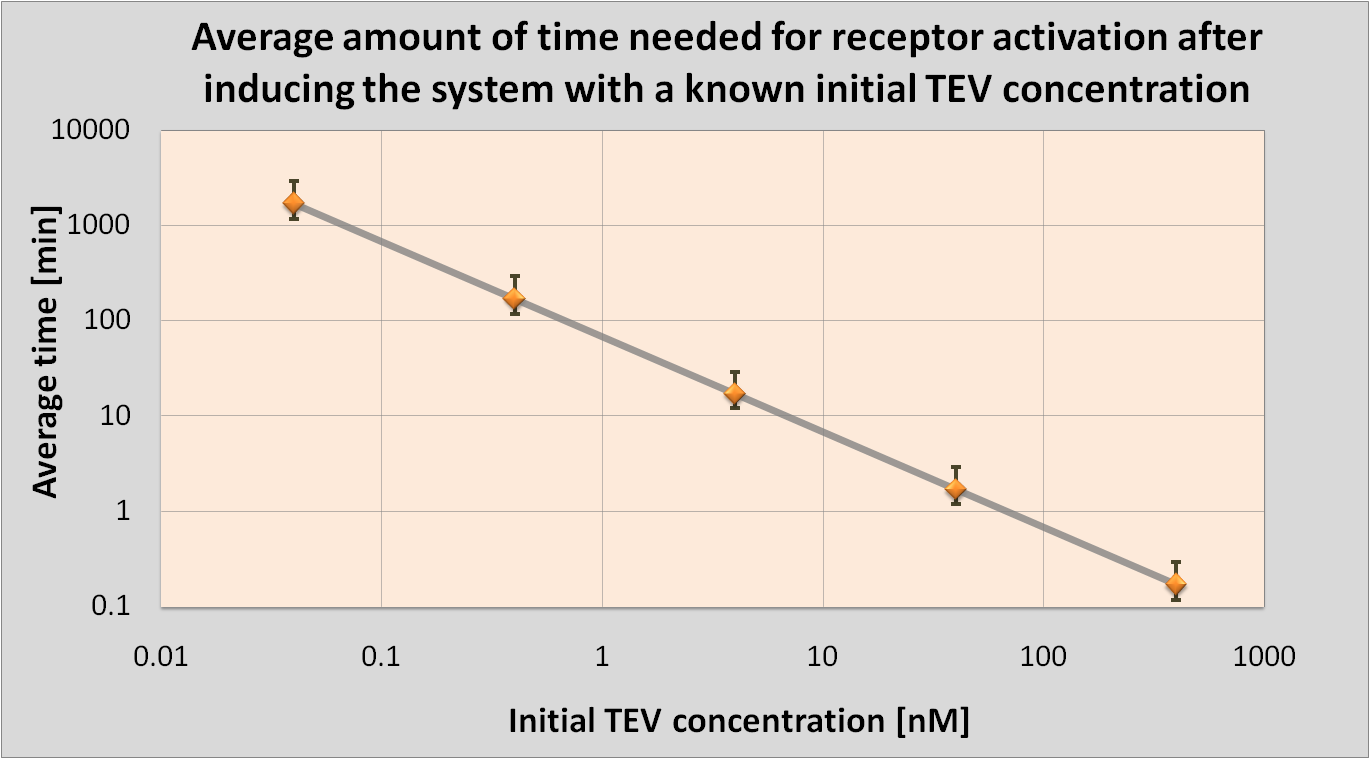
| - | <li>We determined initial TEV protease concentrations which would result in the optimal activation of the receptor. This optimal activation would happen within 1.5 minutes after elastases | + | <li>We determined initial TEV protease concentrations which would result in the optimal activation of the receptor. This optimal activation would happen within 1.5 minutes after elastases come into contact with our cell.</li> |
</ol> | </ol> | ||
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
{| style="background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | {| style="background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
|- | |- | ||
| - | |[[Image:IC_AIP_threshold.png| | + | |[[Image:IC_AIP_threshold.png|600px]] |
|- | |- | ||
| - | | | + | |Notice log-log scale. |
|} | |} | ||
</div> | </div> | ||
|- | |- | ||
| - | |<b> | + | |<b>Signaling Model</b><br/> |
| - | Even though our model of the | + | Even though our model of the signaling module is more simplistic than the real life situation, it provided very important results. We were able to determine under which conditions the signaling pathway would be working and could obtain the major constraints of our system. These constraints are that the necessary concentrations for ComD and AIP are reached before signal transduction is started. |
| + | |||
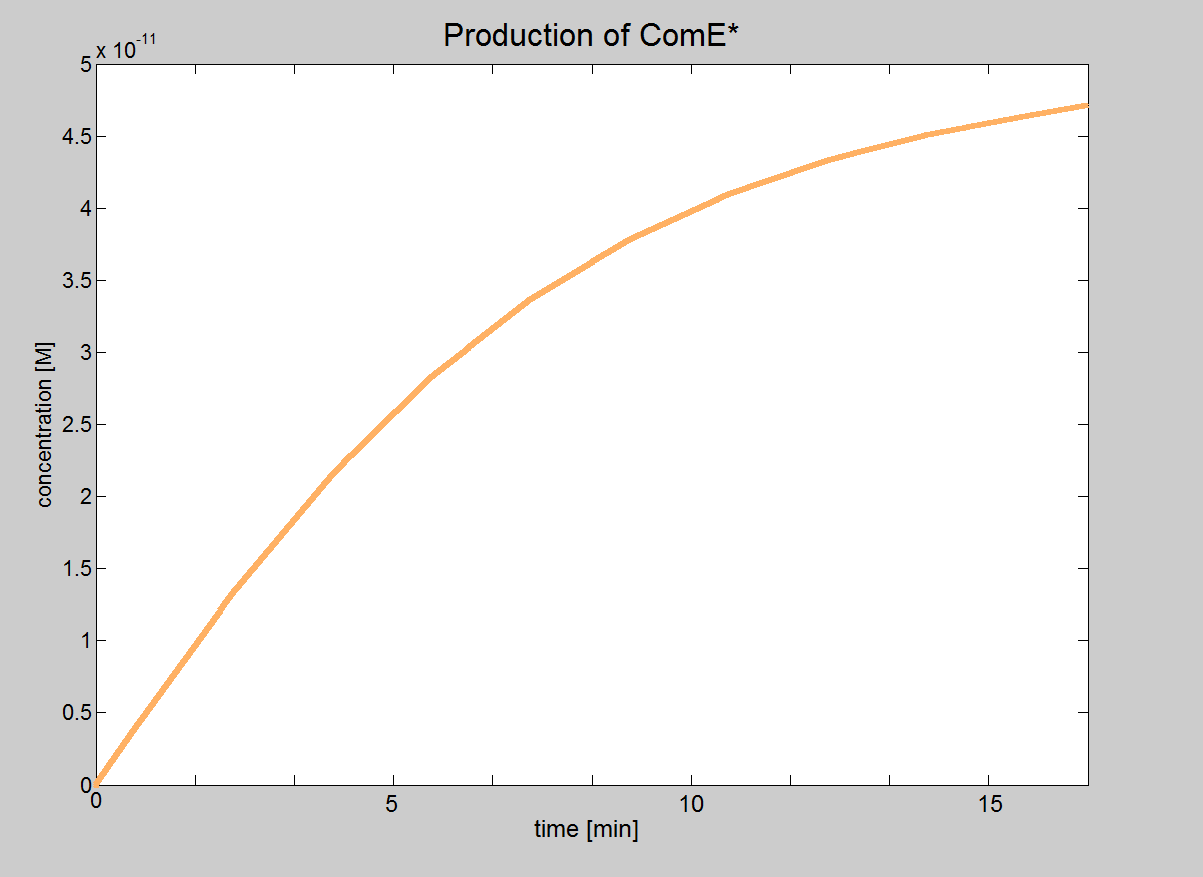
| + | In this model, we will treat phosphorylated ComE (referred to as ComE*) as our "output". Phosphorylated ComE is a transcription factor, which will bind to the DNA to enable transcription. | ||
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
| Line 52: | Line 63: | ||
|[[Image:IC_Signalling_Results1.png|450px]] | |[[Image:IC_Signalling_Results1.png|450px]] | ||
|- | |- | ||
| - | |Graph showing how | + | |Graph showing how the concentration of the transcription |
| + | |- | ||
| + | |factor (phosphorylated ComE) eventually reaches the value 5×10<sup>-11</sup>M. | ||
|} | |} | ||
</div> | </div> | ||
| Line 58: | Line 71: | ||
|<b>Fast Response Model</b><br/> | |<b>Fast Response Model</b><br/> | ||
<ol> | <ol> | ||
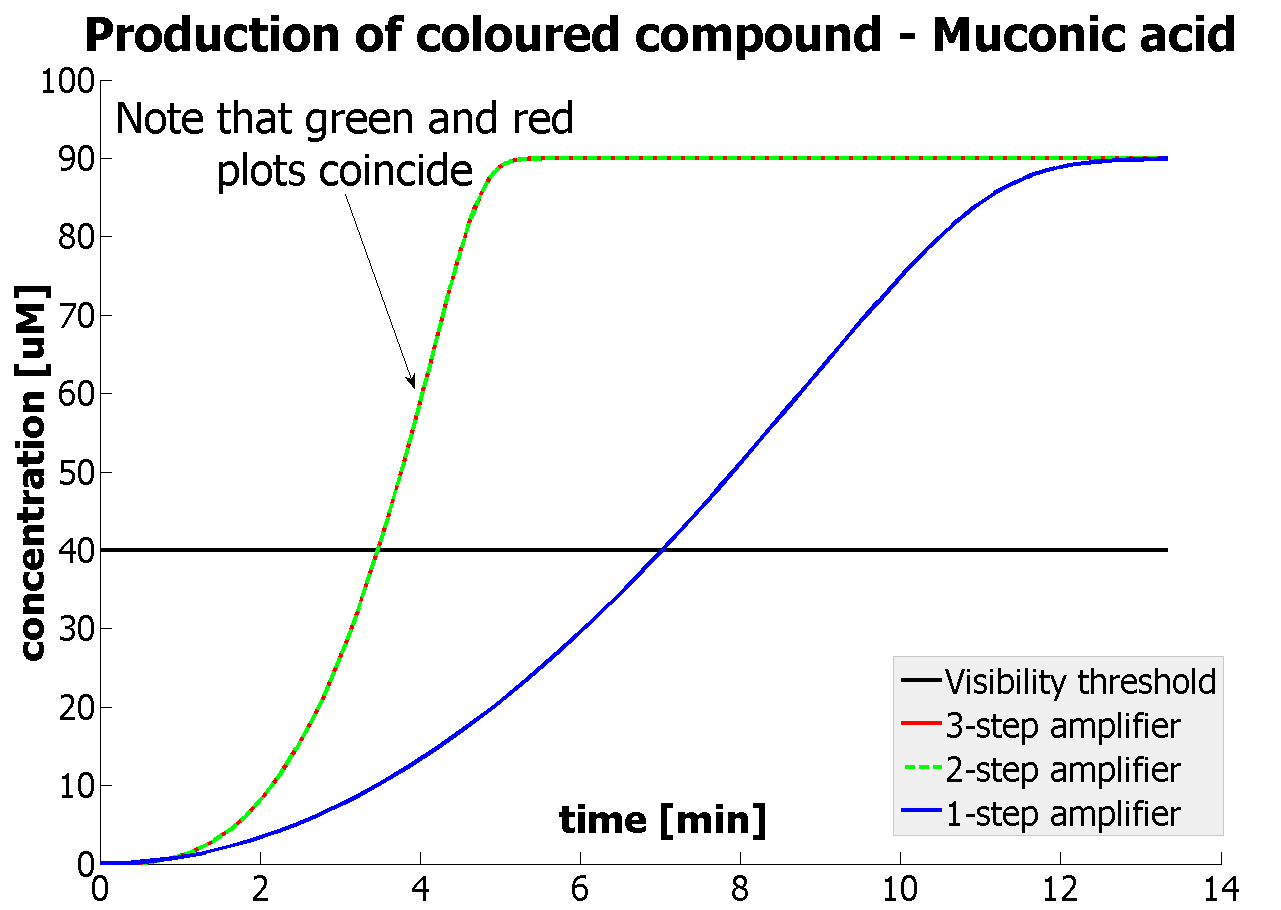
| - | <li>It was shown that our amplification systems easily outperform the simple production system. | + | <li>It was shown that our amplification systems easily outperform the simple production system. Our system can respond within several minutes rather than several hours, which is the case for many currently used reporters. |
<li>It was concluded that there is no advantage of 3-step amplification over 2-step amplification. Therefore, the design of a 3-step amplifier was abandoned.</li> | <li>It was concluded that there is no advantage of 3-step amplification over 2-step amplification. Therefore, the design of a 3-step amplifier was abandoned.</li> | ||
<li>The results concerning the 2-step amplification module were not totally conclusive. It could not be firmly decided whether 2-step amplification is going to perform better than 1-step amplification. This is because several of the parameters that 2- and 1-step amplifiers are sensitive to could not be determined with certainty. Two parameters have been recognised as crucial and decisive. These are the protein production rates and the catalytic constants of the enzymes.</li> | <li>The results concerning the 2-step amplification module were not totally conclusive. It could not be firmly decided whether 2-step amplification is going to perform better than 1-step amplification. This is because several of the parameters that 2- and 1-step amplifiers are sensitive to could not be determined with certainty. Two parameters have been recognised as crucial and decisive. These are the protein production rates and the catalytic constants of the enzymes.</li> | ||
| - | <li>Hence, the conditions for effective amplification were determined.</li> | + | <li>Hence, the conditions for effective amplification were determined (more details are revealed in the Fast Response Model [[Team:Imperial_College_London/Modelling/Output/Results_and_Conclusion|Results]] page).</li> |
</ol> | </ol> | ||
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
| - | {| style="background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | + | {| style="width:454px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; |
|- | |- | ||
|[[Image:Output_model.png|450px]] | |[[Image:Output_model.png|450px]] | ||
|- | |- | ||
| - | |Graph showing how our 2-step | + | |Graph showing how our 2-step amplification system outperforms 1-step amplification. Note that time=0 corresponds to when the first transcription happens upon binding of the transcription factor to DNA. |
|} | |} | ||
</div> | </div> | ||
| Line 80: | Line 93: | ||
|<b>Detection Model</b><br/> | |<b>Detection Model</b><br/> | ||
Goals: | Goals: | ||
| - | <ol><p>The aim of this model is to determine the concentration of Schistosoma elastase or TEV protease that should be added to the bacteria in order to trigger a response | + | <ol><p>The aim of this model is to determine the concentration of Schistosoma elastase or TEV protease that should be added to the bacteria in order to trigger a response.</p> |
| - | <p>It was also attempted to model how long it takes for the protease or elastase to cleave the required amount of peptides.</p> | + | <p>It was also attempted to model how long it takes for the protease or elastase to cleave the required amount of peptides to activate receptors.</p> |
</ol> | </ol> | ||
Elements of the system: | Elements of the system: | ||
| - | <ol><li>The surface protein consists of a cell wall binding domain, linker | + | <ol><li>The surface protein consists of a cell wall binding domain, a linker and AIP (Auto Inducing Peptide)</li> |
| - | <li>Schistosoma elastase (this is the enzyme released by the parasite) cleaves AIP from the cell wall binding domain at the linker site. In the laboratory we used TEV protease as we could not obtain the Schistosoma elastase.</li> | + | <li>Schistosoma elastase (this is the enzyme released by the parasite) cleaves AIP from the cell wall binding domain at the linker site. In the laboratory, we used TEV protease as we could not obtain the Schistosoma elastase.</li> |
<li>The ComD receptor is activated (i.e. AIP concentration is high enough).</li> | <li>The ComD receptor is activated (i.e. AIP concentration is high enough).</li> | ||
</ol> | </ol> | ||
| Line 93: | Line 106: | ||
<ol> | <ol> | ||
<li>The chemical and enzymatic reactions are modelled according to the Law of Mass Action.</li> | <li>The chemical and enzymatic reactions are modelled according to the Law of Mass Action.</li> | ||
| - | <li>Our model assumes that the modelled system is inert within the bacterial | + | <li>Our model assumes that the modelled system is inert within the bacterial cell or that reactions with other species within the bacterium is negligible. For example, the TEV protease is not supposed to cleave other molecules due to its specifity.</li> |
<li>Due to our carefully chosen cell concentrations, the diffusion of free AIPs could be neglected. However, this restricts the model to the considered cell concentrations only.</li> | <li>Due to our carefully chosen cell concentrations, the diffusion of free AIPs could be neglected. However, this restricts the model to the considered cell concentrations only.</li> | ||
<li>The threshold for receptor activation was defined by one specific value as opposed to considering intermediate states between fully "off" and "on".</li> | <li>The threshold for receptor activation was defined by one specific value as opposed to considering intermediate states between fully "off" and "on".</li> | ||
</ol> | </ol> | ||
|- | |- | ||
| - | |<b> | + | |<b>Signaling Model</b><br/> |
Goals: | Goals: | ||
| - | The aim of this model is to determine under which conditions the | + | The aim of this model is to determine under which conditions the signaling transduction will happen in our bacteria. |
Elements of the system: | Elements of the system: | ||
<ol> | <ol> | ||
| - | <li>The | + | <li>The signaling model consists of ComD and ComE, which are expressed by our bacteria, as well as AIP and Phosphate. These species are all assumed to be present in the cell at a sufficient concentration.</li> |
<li>The ComD receptor is activated by the AIP. This triggeres phosphorylation of the ComD receptor. The Phosphate group of the ComD receptor then binds to ComE. The phosphorylated ComE binds to the DNA and acts as a transcription factor.</li> | <li>The ComD receptor is activated by the AIP. This triggeres phosphorylation of the ComD receptor. The Phosphate group of the ComD receptor then binds to ComE. The phosphorylated ComE binds to the DNA and acts as a transcription factor.</li> | ||
</ol> | </ol> | ||
| Line 120: | Line 133: | ||
</ol> | </ol> | ||
Elements of the system: | Elements of the system: | ||
| - | <ol><li>Dioxygenase (<i>blue on the diagrams below</i>) is an enzyme that acts on catechol to produce a yellow output. In most of our models dioxygenase was treated as an output because it was found that active dioxygenase acting on catechol produces the coloured output within a split second.</li> | + | <ol><li>Dioxygenase (<i>blue on the diagrams below</i>) is an enzyme that acts on catechol to produce a yellow output. In most of our models, dioxygenase was treated as an output because it was found that active dioxygenase acting on catechol produces the coloured output within a split second.</li> |
| - | <li>GFP-Dioxygenase fusion protein (<i>GFP is shown green on the diagrams</i>). Dioxygenase joined by the linker | + | <li>GFP-Dioxygenase fusion protein (<i>GFP is shown green on the diagrams</i>). Dioxygenase, which is joined to GFP by the linker, was assumed to be inactive.</li> |
<li>TEV protease (<i>pink on the diagrams below</i>) has the ability to cleave the GFP-Dioxygenase fusion protein, hence, it activates dioxygenase</li> | <li>TEV protease (<i>pink on the diagrams below</i>) has the ability to cleave the GFP-Dioxygenase fusion protein, hence, it activates dioxygenase</li> | ||
<li>Split TEV protease (<i>purple on the diagrams below</i>) is an inactive split form of TEV mounted on coiled coils. It can be activated again by coiled coils being cleaved by another active TEV.</li> | <li>Split TEV protease (<i>purple on the diagrams below</i>) is an inactive split form of TEV mounted on coiled coils. It can be activated again by coiled coils being cleaved by another active TEV.</li> | ||
| Line 131: | Line 144: | ||
|[[Image:Simple_Production.png|700px]] | |[[Image:Simple_Production.png|700px]] | ||
|- | |- | ||
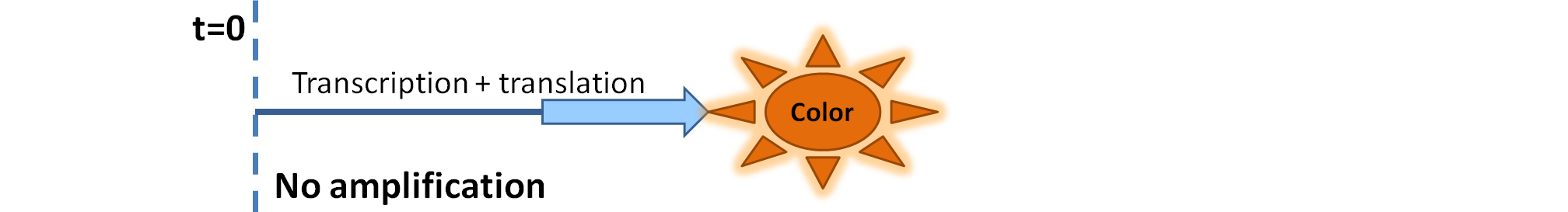
| - | |Simple production upon activation of arbitrary colour output by transcription and translation indicated by the <i>blue arrow</i>. | + | |Simple production upon activation of an arbitrary colour output by transcription and translation indicated by the <i>blue arrow</i>. |
|- | |- | ||
|[[Image:1-step_amplification.png|700px]] | |[[Image:1-step_amplification.png|700px]] | ||
|- | |- | ||
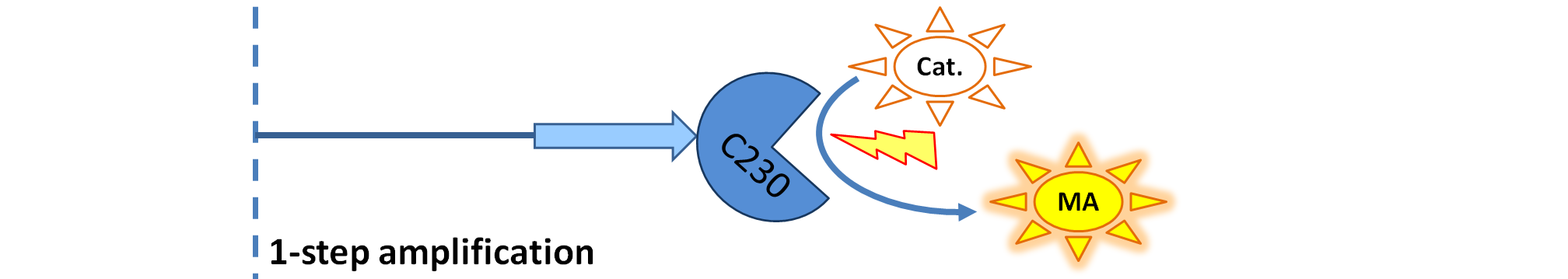
| - | | Dioxygenase (C230) is simply produced. Upon activation at time t=0, it acts on catechol (cat.) to produce yellow output - muconic acid. Catechol is not shown to be produced by cell as it is added | + | | Dioxygenase (C230) is simply produced. Upon activation at time t=0, it acts on catechol (cat.) to produce the yellow output - muconic acid. Catechol is not shown to be produced by the cell as it is added manually at an arbitrary time. |
|- | |- | ||
|[[Image:2-step_amplification.png|700px]] | |[[Image:2-step_amplification.png|700px]] | ||
|- | |- | ||
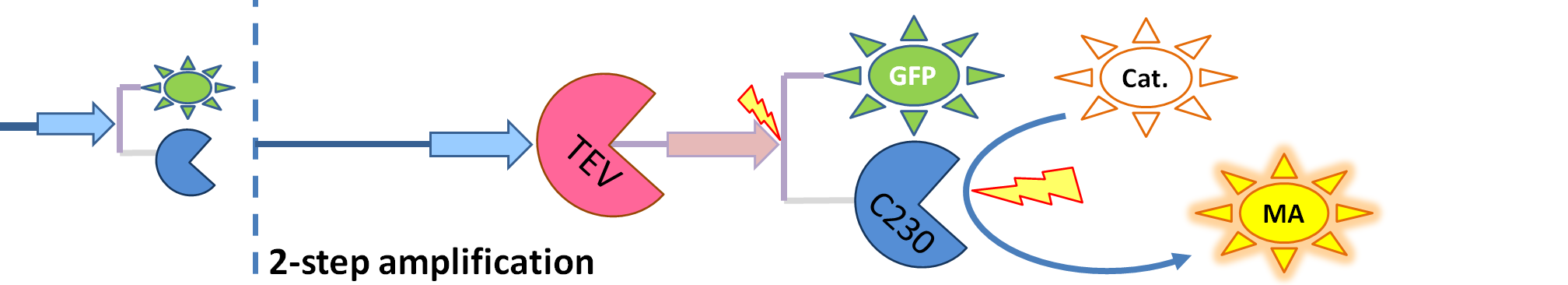
| - | | The species that are shown in front of vertical line which indicates beginning of experiment | + | | The species that are shown in front of the vertical line (which indicates the beginning of the experiment) indicate that they have been accumulated beforehand in the cell. TEV protease activates inactive dioxygenase which acts on catechol to produce colour. |
|- | |- | ||
|[[Image:3-step_amplification.png|700px]] | |[[Image:3-step_amplification.png|700px]] | ||
|- | |- | ||
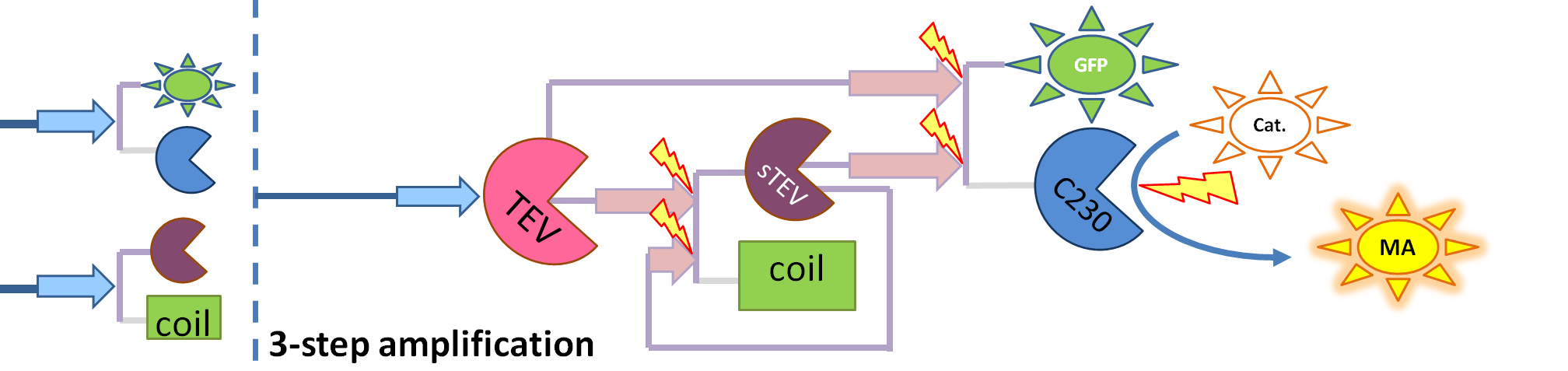
| - | | This diagram introduces inactive split TEV protease attached to a coiled-coil as the third amplification step. Both inactive compounds have active | + | | This diagram introduces inactive split TEV protease attached to a coiled-coil as the third amplification step. Both inactive compounds have active sites for TEV to activate them, which results in multiple possibilities of action. |
|} | |} | ||
</div> | </div> | ||
| Line 150: | Line 163: | ||
<ol> | <ol> | ||
<li>The chemical and enzymatic reactions are modelled according to the Law of Mass Action.</li> | <li>The chemical and enzymatic reactions are modelled according to the Law of Mass Action.</li> | ||
| - | <li>Our model assumes that the modelled system is inert within the bacterial | + | <li>Our model assumes that the modelled system is inert within the bacterial cell or that reactions with other species within the bacterium is negligible. For example, the TEV protease is not supposed to cleave other molecules due to its specifity.</li> |
</ol> | </ol> | ||
|} | |} | ||
Latest revision as of 03:59, 28 October 2010
| Modelling | Overview | Detection Model | Signaling Model | Fast Response Model | Interactions |
| A major part of the project consisted of modelling each module. This enabled us to decide which ideas we should implement. Look at the Fast Response page for a great example of how modelling has made a major impact on our design! | |
| Introduction to modelling | ||||||
| During the design of our construct three major questions arose which could be answered by computer modelling:
We came up with a novel idea of detecting organisms that we do not have a specific receptor for. In our particular example, the protease of Schistosoma is meant to cleave a protein displayed on the bacteria's cell wall (the pink rectangle on the diagram below). The cleaved peptide is recognized by the receptor which would then activate the colour expression. This solution raised questions about the risk of false positive or whether there are any chances for ComD receptors to be activated in the diluted environment. Modelling of this module would enable us to answer these questions.
We decided to use the ComCDE signaling pathway from S.pneumoniae and so questions arose on whether it would work appropriately in B.subtilis. We modelled this system to make sure that the signalling pathway would be working as anticipated (the green rectangle on the diagram below).
We came up with the idea of using the amplification of a colour output, which would show within minutes of the stimulus being added (the blue rectangle on the diagram below). The question that arose was whether amplification will actually perform better than simple production (i.e. transcription and translation) in the cellular environment. Furthermore, we had difficulties deciding whether we should design the amplification module to consist of 1,2 or even more amplification steps. These issues were quite difficult to answer, so we decided to employ modelling. |
| Results & Conclusions |
Detection Model
|
| Signaling Model Even though our model of the signaling module is more simplistic than the real life situation, it provided very important results. We were able to determine under which conditions the signaling pathway would be working and could obtain the major constraints of our system. These constraints are that the necessary concentrations for ComD and AIP are reached before signal transduction is started. In this model, we will treat phosphorylated ComE (referred to as ComE*) as our "output". Phosphorylated ComE is a transcription factor, which will bind to the DNA to enable transcription. |
Fast Response Model
|
 "
"