Team:Heidelberg/Project/Capsid Shuffling/Homology Based
From 2010.igem.org
(→Introduction) |
(→Results and Discussion) |
||
| (20 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
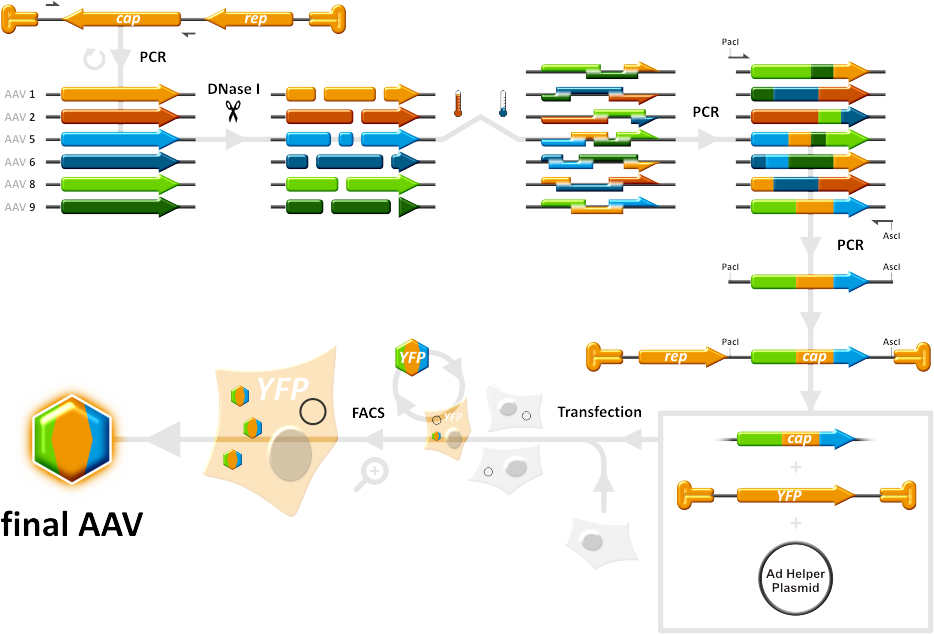
Our main goal was to produce a library of Adeno-associated viruses that infect cells with different specificities and efficiencies, due to differing capsid composition, and select from this library the best AAVs that are highly specific. Since it is not the case that viruses that work well in vitro will work as good in vivo, we had the aim of testing our candidates in mice as well. To achieve this, the capsid genes from AAV serotypes 1,2,5,6,8 and 9 were shuffled between each other in a primerless PCR reaction that relies on the close homology between the different cap genes. Those capsid genes were first digested with DNase I, then pooled together and allowed to anneal to each other. A second PCR was done then to amplify the shuffled cap gene fragments that were generated in the first PCR while introducing AscI and PacI restriction sites, which are used to clone the cap genes into a helper vector. AAVs that were then produced in HEK293 cells, which were transfected with the cap-gene-ITR construct and another Adeno helper construct, made up the viral library that was subjected to selection pressure in target cells. In this manner, only AAVs have the most fitting caspsids will successfully infect target cells and produce more viral particles with the help of Adeno virus. By taking the viruses that survive one selection round and applying them to the next, we would end up with a group of highly efficient and specific viruses that overcome many of the issues with the wild-type AAVs. | Our main goal was to produce a library of Adeno-associated viruses that infect cells with different specificities and efficiencies, due to differing capsid composition, and select from this library the best AAVs that are highly specific. Since it is not the case that viruses that work well in vitro will work as good in vivo, we had the aim of testing our candidates in mice as well. To achieve this, the capsid genes from AAV serotypes 1,2,5,6,8 and 9 were shuffled between each other in a primerless PCR reaction that relies on the close homology between the different cap genes. Those capsid genes were first digested with DNase I, then pooled together and allowed to anneal to each other. A second PCR was done then to amplify the shuffled cap gene fragments that were generated in the first PCR while introducing AscI and PacI restriction sites, which are used to clone the cap genes into a helper vector. AAVs that were then produced in HEK293 cells, which were transfected with the cap-gene-ITR construct and another Adeno helper construct, made up the viral library that was subjected to selection pressure in target cells. In this manner, only AAVs have the most fitting caspsids will successfully infect target cells and produce more viral particles with the help of Adeno virus. By taking the viruses that survive one selection round and applying them to the next, we would end up with a group of highly efficient and specific viruses that overcome many of the issues with the wild-type AAVs. | ||
| - | + | == Results and Discussion == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | '''Selection in Huh-7 cells:''' | ||
| + | |||
| + | Three selection rounds were carried out on Huh-7 cells. We applied negative selection pressure set onto the speed of infection by washing away the virus after 30 min of infection time. The viral DNA was isolated for each selection round and cloned into a Wilson AAV8 wild-type vector. 12 clones from each selection round were picked, and viruses that contain a YFP construct were produced and used to infect 96-well plates of mouse primary hepatocytes, HeLa and Huh-7 cells. We already have some results for shuffled and selected AAV but they dont represent the whole variety of clones that are still breading inside the incubator and will be tested with facs analysis this week. | ||
| + | |||
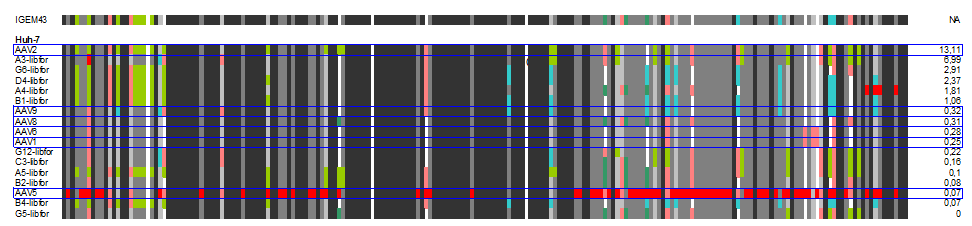
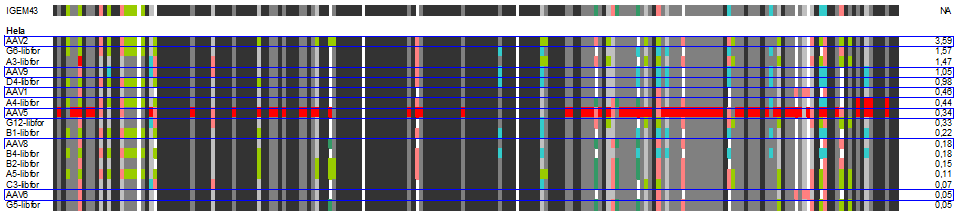
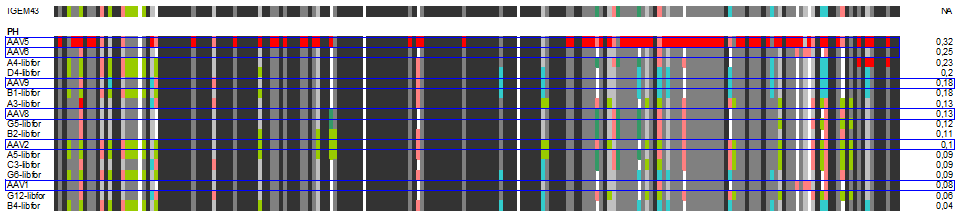
| + | The following alignment plots show alignment results for eleven different clones obtained from supernatant after selection round 1. Since there are no viruses from further selection rounds available yet, the plot does not contain enough samples for displaying the evolutionary (based on selection only) process. The samples were compared to wild types and sorted for infection efficiency in HUH-7 Hep-G2 and primary hepatocytes, respectively, measured in FACS experiments. In addition, the Alignment of the clone 43 used in mouse experiments is added for pattern comparison. | ||
| + | |||
| + | [[Image:huh_clones.gif|600px]] | ||
| + | [[Image:legende.gif|600px]] | ||
| + | [[Image:hela_clones.gif|600px]] | ||
| + | [[Image:legende.gif|600px]] | ||
| + | [[Image:PH_clones.gif|600px]] | ||
| + | [[Image:legende.gif|600px]] | ||
| + | |||
| + | In our alignment plot it can clearly be seen that there are certain patterns that add to the ability of transducing target cells (what. Aims for the experiments of the next week will be the monitoring of kinetics of the selection process as well as finding patterns that might be responsible for expression levels inside the infected cells that seem to be independant of transfection efficiency in some of our FACS measurments. | ||
| + | |||
| + | '''Selection in Mouse Primary Hepatocytes:''' | ||
| + | |||
| + | The fact that mouse primary hepatocytes are particularly hard to infect adds to the selection pressure applied on the AAV library. Two selection rounds were carried out on those cells, and viral DNA was extracted and cloned into the Wilson AAV8 wild-type vector. 20 clones were selected from each selection round and will be sequenced today and tested on Monday next week with a HAAT construct on primary hepatocytes. | ||
| + | |||
| + | '''Randomly picked clones''' | ||
| + | |||
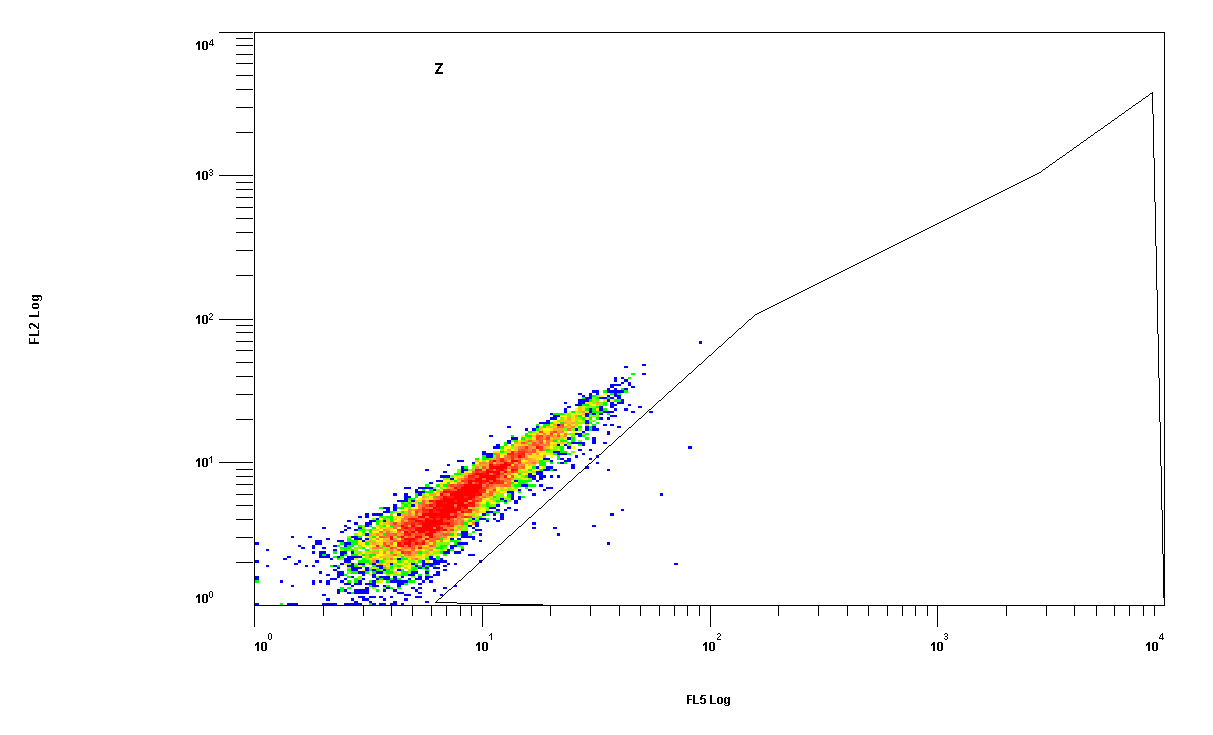
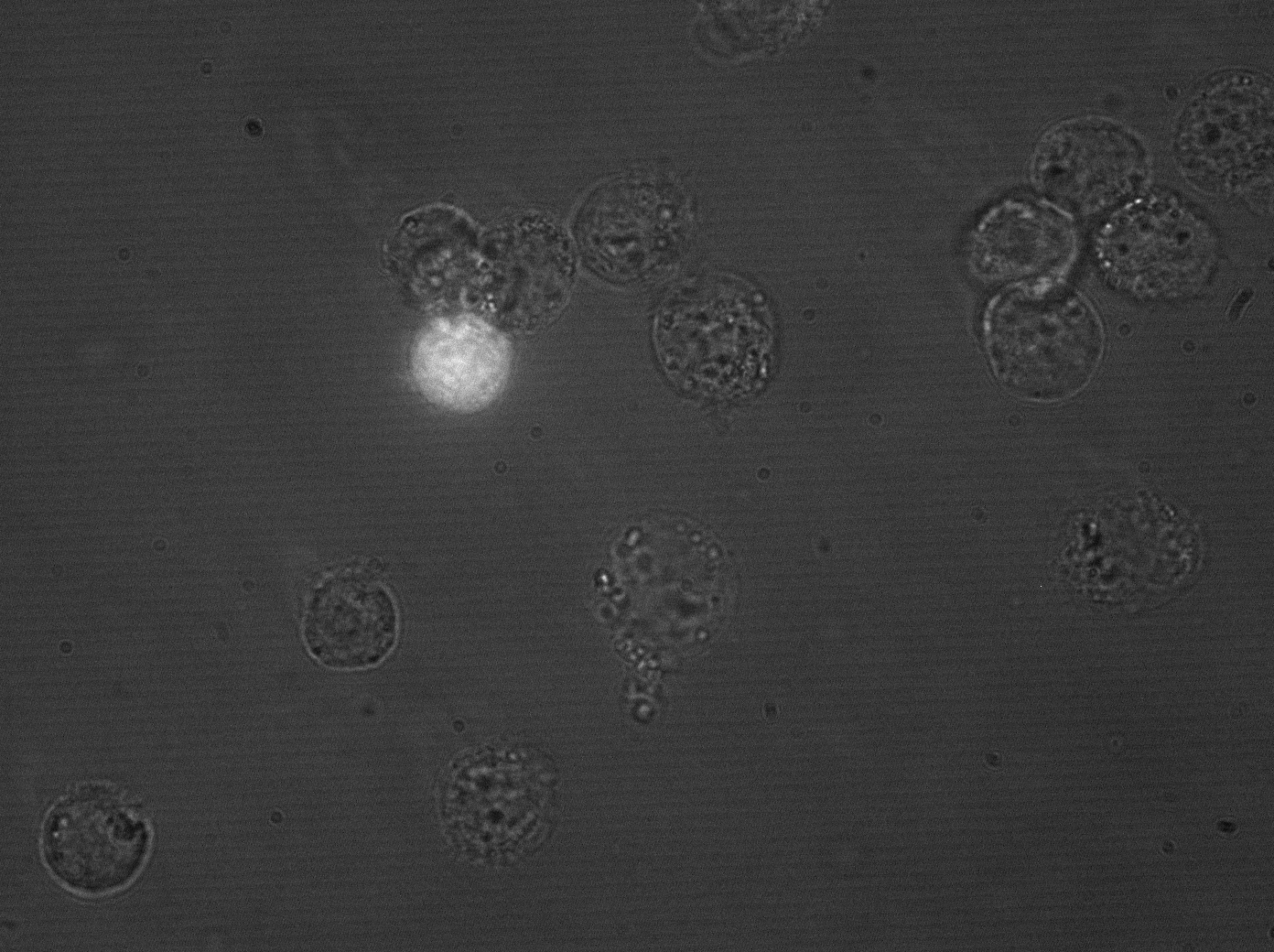
| + | In parallel, we picked 48 randomly selected clones from the AAV library, packaged them with a YFP construct and tested them using flow cytometry on HepG2, Huh-7 and mouse primary hepatocytes. One clone (#43) of those was able to infect both HepG2 and Huh-7 cells, and was thus packaged with a luciferase construct and injected into mice. [https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection Mice experiment] showed a specific targeting into the liver. | ||
| + | |||
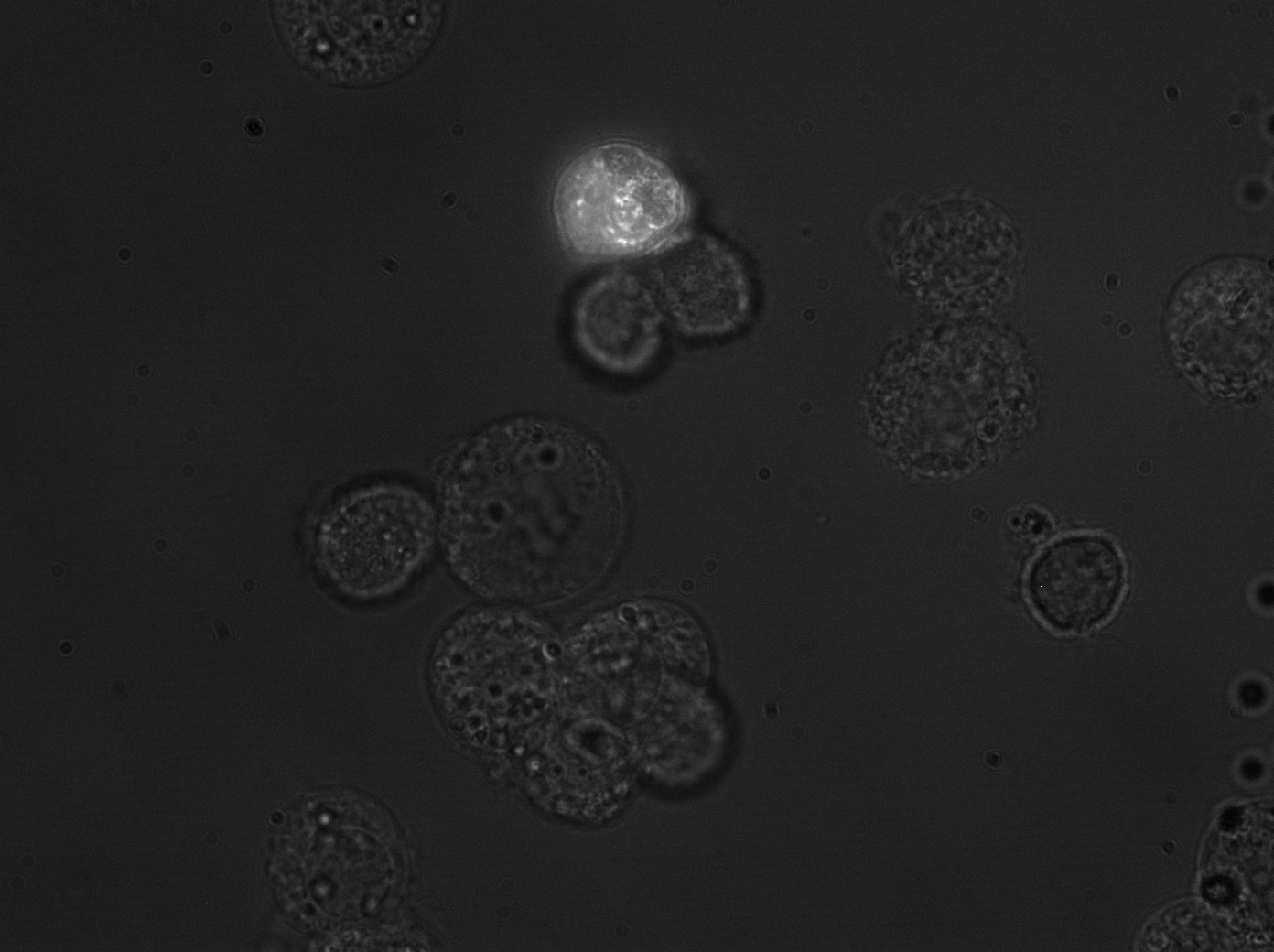
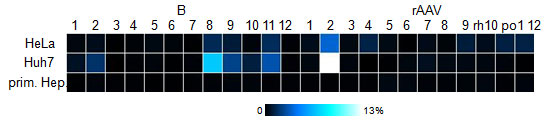
| + | In addition, 12 viruses were packaged with a YFP construct and used to transduce primary hepatocytes, HeLa and Huh-7 cells. Flow cytometry measurements showed that some of those viruses (e.g. number 8) are even more efficient than many of the wildtype serotypes. However, the measurements will be repeated, since not enough time was allowed for the viruses to be produced and for the cells to express sufficient YFP for good measurements. | ||
| + | Sequencing data were used for the alignment of those 12 clones with 10 AAV wildtypes (see above). | ||
| + | |||
| + | |||
| + | [[Image:facs_43_huh.gif|200px|clone number 43, Huh-7 cells]] | ||
| + | |||
| + | [[Image:hepg2_transyfp_2.gif|Clone 43 under microscope, HepG2 cells|300px]] | ||
| + | |||
| + | [[Image:huh7_transyfp1_2.gif|300px]] | ||
| + | |||
| + | [[Image:Flowcyt.jpg|Flow cytometry result for the 12 clones as compared to the 12 wildtypes|300px]] | ||
| - | |||
{{:Team:Heidelberg/Pagemiddle}} | {{:Team:Heidelberg/Pagemiddle}} | ||
| + | [[Image:HomBasShuffling 934px.png|270px]] | ||
{{:Team:Heidelberg/Bottom}} | {{:Team:Heidelberg/Bottom}} | ||
Latest revision as of 03:59, 28 October 2010

|
|
||
 "
"