Team:Heidelberg/Notebook/Material Methods
From 2010.igem.org
(→Buffers) |
|||
| (48 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Double}} |
{{:Team:Heidelberg/Pagetop|home}} | {{:Team:Heidelberg/Pagetop|home}} | ||
=Materials= | =Materials= | ||
| - | == | + | == Kits == |
| - | ==== | + | <center> |
| - | + | {| class="wikitable sortable" border="0" style="text-align: left" | |
| + | |-bgcolor=#cccccc | ||
| + | |+ align="top, left"|'''table 1''': Used kits and referring suppliers. | ||
| + | | Kits||Supplier||Catalog Number | ||
| + | |- | ||
| + | |HiSpeed® Plasmid Maxi Kit (25)||QIAGEN||12663 | ||
| + | |- | ||
| + | |CompactPrep Plasmid Maxi Kit (25)||QIAGEN||12763 | ||
| + | |- | ||
| + | |RNeasy® Mini Kit (25)||QIAGEN||74106 | ||
| + | |- | ||
| + | |QIAquick® Miniprep Kit (250)||QIAGEN||27106 | ||
| + | |- | ||
| + | |QIAquick® Gel Extraction Kit (250)||QIAGEN||28706 | ||
| + | |- | ||
| + | |QIAquick® Nucleotid Removal Kit (250)||QIAGEN||28306 | ||
| + | |- | ||
| + | |QIAquick® PCR Purification Kit (250)||QIAGEN||28106 | ||
| + | |- | ||
| + | |GeneMorph® II EZC Clone Domain Mutagenesis Kit||Stratagene||200552-5 | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| - | + | == Marker == | |
| + | <center> | ||
| + | {| class="wikitable sortable" border="0" style="text-align: left" | ||
| + | |-bgcolor=#cccccc | ||
| + | |+ align="top, left"|'''table 2''': Used DNA Ladder for Gel Electrophoresis. | ||
| + | | Name||Supplier||Range | ||
| + | |- | ||
| + | |GeneRuler™ High Range DNA Ladder||Fermentas||10,171-48,502 bp | ||
| + | |- | ||
| + | |GeneRuler™ 1 kb Plus DNA Ladder||Fermentas||75-20,000 bp | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| - | + | == Enzymes == | |
| + | <center> | ||
| + | {| class="wikitable sortable" border="0" style="text-align: left" | ||
| + | |-bgcolor=#cccccc | ||
| + | |+ align="top, left"|'''table 3''': Used NEB restriction enzymes | ||
| + | | Name||Number/ID||NEB buffer1||NEB buffer2||NEB buffer3||NEB buffer4||BSA | ||
| + | |- | ||
| + | |AflII||27||50%||100%||25%||100%||x | ||
| + | |- | ||
| + | |AgeI||1||100%||50%||10%||75% | ||
| + | |- | ||
| + | |ApaI||-||25%||50%||0%||100%||x | ||
| + | |- | ||
| + | |AseI||not||75%||100%||not|| | ||
| + | |- | ||
| + | |AvrII||100%||100%||50%||100%|| | ||
| + | |- | ||
| + | |BamHI||2||75%||100%||100%||100%||x | ||
| + | |- | ||
| + | |BamHI||-||75%||100%||100%||100%||x | ||
| + | |- | ||
| + | |BclI||-||50%||100%||100%||75%|| | ||
| + | |- | ||
| + | |BglI||-||50%||75%||100%||50%|| | ||
| + | |- | ||
| + | |BglII||13|| 10%|| 75%|| 100%|| 10%|| | ||
| + | |- | ||
| + | |BsrGI||-|| 25%|| 100%|| 10%|| 100%||x | ||
| + | |- | ||
| + | |DpnI ||6|| 100%|| 100%|| 75%|| 100%|| | ||
| + | |- | ||
| + | |DpnI ||-|| 100%|| 100%|| 75%|| 100%|| | ||
| + | |- | ||
| + | |EcoRI ||-|| 100%|| 100%|| 100%|| 100%|| | ||
| + | |- | ||
| + | |EcoRV ||-|| 50%|| 75%|| 100%|| 50%|| x | ||
| + | |- | ||
| + | |FseI ||-|| 100%|| 75%|| 0%|| 100%|| | ||
| + | |- | ||
| + | |HindIII||-|| 10 50%|| 100%|| 10%|| 50%|| | ||
| + | |- | ||
| + | |HindIII||-||50%|| 100%|| 10%|| 50%|| | ||
| + | |- | ||
| + | |KpnI ||11|| 100%|| 75%|| 0%|| 50%|| x | ||
| + | |- | ||
| + | |MfeI-HF™||-|| 75%|| 50%|| 10%|| 100%|| | ||
| + | |- | ||
| + | |NarI ||26|| 100%|| 75%|| 75%|| 100%|| | ||
| + | |- | ||
| + | |NcoI ||12|| 100%|| 100%|| 100%|| 100%|| | ||
| + | |- | ||
| + | |NdeI ||-|| 75%|| 100%|| 75%|| 100%|| | ||
| + | |- | ||
| + | |NheI ||-|| 100%|| 100%|| 10%|| 100%|| x | ||
| + | |- | ||
| + | |NotI ||14|| 0%|| 50%|| 100%|| 25%|| | ||
| + | |- | ||
| + | |NsiI ||-|| 10%|| 75%|| 100%|| 25%|| | ||
| + | |- | ||
| + | |PciI ||-|| 50%|| 75%|| 100%|| 50%|| x | ||
| + | |- | ||
| + | |PstI ||15|| 75%|| 75%|| 100%|| 50%|| | ||
| + | |- | ||
| + | |SacI ||16|| 100%|| 50%|| 10%|| 100%|| | ||
| + | |- | ||
| + | |ScaI ||18|| not||not||100%||not|| | ||
| + | |- | ||
| + | |SfcI ||19|| 75%|| 50%|| 10%|| 100%|| x | ||
| + | |- | ||
| + | |SfcI||-||75%|| 50%|| 10%|| 100%|| x | ||
| + | |- | ||
| + | |SpeI ||-|| 75%|| 100%|| 25%|| 100%|| x | ||
| + | |- | ||
| + | |SphI ||-|| 100%|| 100%|| 50%|| 100%|| | ||
| + | |- | ||
| + | |SspI ||-|| 50%|| 100%|| 50%|| 50%|| | ||
| + | |- | ||
| + | |XbaI ||23 ||0%|| 100%|| 75%|| 100%|| x | ||
| + | |- | ||
| + | |XbaI ||56|| 0%|| 100%|| 75%|| 100%|| x | ||
| + | |- | ||
| + | |XhoI||24|| 75%|| 100%|| 100%|| 100%|| x | ||
| + | |- | ||
| + | |XmaI||25|| 25%|| 50%|| 0%|| 100%|| x | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| + | NEBuffers are color-coded (NEBuffer 1-yellow, NEBuffer 2-blue, | ||
| + | NEBuffer 3-red, NEBuffer 4-green) and supplied as 10X | ||
| - | *''' | + | == Bacteria == |
| + | <center> | ||
| + | {| class="wikitable sortable" border="0" style="text-align: left" | ||
| + | |-bgcolor=#cccccc | ||
| + | |+ align="top, left"|'''table 4''': Used <i>E. coli</i> Strains. | ||
| + | | Name||Purpose||Reference | ||
| + | |- | ||
| + | |DH5α||Amplification of Plasmids||Invitrogen | ||
| + | |- | ||
| + | |SCS110||Amplification of unmethylated Plasmids||Stratagene | ||
| + | |- | ||
| + | |Top10||Amplification of Plasmids||Invitrogen | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| + | === Media and Antibiotics === | ||
| + | * '''LB''' (10 g tryptone, 5 g yeast extract, 10 g sodium chloride, ad 1 L ddH<sub>2</sub>O) | ||
| + | <center> | ||
| + | {| class="wikitable sortable" border="0" style="text-align: left" | ||
| + | |-bgcolor=#cccccc | ||
| + | |+ align="top, left"|'''table 5''': Used Antibiotics. | ||
| + | ! Antibiotic||Final Concentration|| Solvent | ||
| + | |- | ||
| + | |Ampicillin||100 µg/ml||ddH<sub>2</sub>O | ||
| + | |- | ||
| + | |Chloramphenicol||25 µg/ml||100% Ethanol | ||
| + | |- | ||
| + | |Kanamycin||50 µg/ml||ddH<sub>2</sub>O | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| - | |||
| - | |||
| + | =Methods= | ||
| + | ==Cloning== | ||
| - | + | ===Agarose Gel Electrophoresis=== | |
| - | + | Agarose flat-bed gels in various concentrations (0.6–2% agarose in 0.5 x TAE buffer) and sizes were run to separate DNA fragments in an electrical field (10–20 V/cm) for analytical or preparative use. The desired amount of agarose was boiled in 1 x TAE buffer until it was completely dissolved. After it cooled down to approximately 60°C, ethidium bromide (EtBr) solution (0.5 μg/ml final concentration) was added to the liquid agar, which was then poured in a flat-bed tray with combs. As soon as the agarose solidified, the Running buffer (0.5 x TAE buffer) was added before the DNA in the loading buffer was loaded into the wells and separated electrophoretically. Ethidium bromide intercalates with the DNA’s GC ntss resulting in DNA-EtBr-complex that emits visible light. Therefore, the DNA fragments could be detected on a UV-light tray at 265 nm. | |
| - | + | ||
| - | + | ||
| - | + | ===Colony PCR=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Colony PCRs were performed using Fermentas PCR Master Mix (2x), containing ''Taq'' DNA Polymerase.<br/> | |
| - | + | Bacterial colonies were picked form an LB/Agar-plate and either resuspended in PCR reaction mix and subsequently used to inoculate LB-medium with appropriate antibiotic, or resuspended in water. The water was then taken as "template" for PCR and inoculation of Minepreps.<br/> | |
| + | The PCR was performed using 20 pmol of forward and reverse primer in a total volume of 20 µL.<br/> | ||
| + | PCR conditions were set to recommendations made by Fermentas: | ||
| - | === | + | {| border="1" cellpadding="3" style="border:solid 1px #AAAAAA; border-collapse:collapse; background-color:#F9F9F9; empty-cells:show;" |
| - | + | ||
| - | + | !Temp !! Time !! | |
| - | === | + | |- |
| - | + | |95°||03:00|| | |
| + | |- | ||
| + | |style="border-top:solid 1px #000000;"|94°||style="border-top:solid 1px #000000;border-right:solid 1px #000000;"|00:30||rowspan="3"|x 30 cycles | ||
| + | |- | ||
| + | |45°'*'||style="border-right:solid 1px #000000;"|00:30 | ||
| + | |- | ||
| + | |style="border-bottom:solid 1px #000000;"|72°||style="border-bottom:solid 1px #000000;border-right:solid 1px #000000;"|01:00 /kb | ||
| + | |- | ||
| + | |72°||10:00|| | ||
| + | |- | ||
| + | |4°||8|| | ||
| + | |} | ||
| + | |||
| + | '*'with 45°C at a minimum of primer annealing temp | ||
===Gel extraction=== | ===Gel extraction=== | ||
After gel electrophoresis the digested vector and insert have to be purified from the gel. With the help of a UV lamp, the bands were quickly excised from the gel without exposing the DNA too long to UV light. Afterwards the DNA was purified with the QIAquick Gel extraction kit. Three volumes of buffer QG were added to one volume of gel. The gel fragment was dissolved by incubation for 10 min at 50°C. Afterwards one volume of 100% isopropanol was added. The solution was applied on a QIAquick spin column after this has been placed into a provided 2 ml collection tube. By centrifugation for 1 min at 13.000 rpm the DNA was bound to the column. The flow-through was discarded and the column was placed in the same collection tube. To remove all traces of agarose from the column, 500 µl of wash buffer QC was added followed by centrifugation for 1 min at 13.000 rpm. The flow-through was discarded and the column was washed with 750 µl of buffer PE for 1 min at 13.000 rpm. Afterwards the flow-through was discarded. An additional centrifugation for 1 min at 13.000 rpm helped to remove the residual ethanol. The column was placed into a new 1.5 ml microcentrifuge tube and it was eluted with 30 µl of ddH2O. | After gel electrophoresis the digested vector and insert have to be purified from the gel. With the help of a UV lamp, the bands were quickly excised from the gel without exposing the DNA too long to UV light. Afterwards the DNA was purified with the QIAquick Gel extraction kit. Three volumes of buffer QG were added to one volume of gel. The gel fragment was dissolved by incubation for 10 min at 50°C. Afterwards one volume of 100% isopropanol was added. The solution was applied on a QIAquick spin column after this has been placed into a provided 2 ml collection tube. By centrifugation for 1 min at 13.000 rpm the DNA was bound to the column. The flow-through was discarded and the column was placed in the same collection tube. To remove all traces of agarose from the column, 500 µl of wash buffer QC was added followed by centrifugation for 1 min at 13.000 rpm. The flow-through was discarded and the column was washed with 750 µl of buffer PE for 1 min at 13.000 rpm. Afterwards the flow-through was discarded. An additional centrifugation for 1 min at 13.000 rpm helped to remove the residual ethanol. The column was placed into a new 1.5 ml microcentrifuge tube and it was eluted with 30 µl of ddH2O. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
===Large scale preparation of plasmid DNA=== | ===Large scale preparation of plasmid DNA=== | ||
150 ml LB-Medium with 150 µl ampicillin was inoculated with 50 µl of bacteria culture which grew overnight on a shaker at 37°C. The plasmid DNA was isolated using QiAprep Spin MAxiprep kit from Qiagen and the protocol was followed. The overnight culture was centrifuged for 20 min at 4000 rpm at 4°C using an SLA 1500 Rotor. Afterwards the LB-medium was discarded and the pellet was homogeneously resuspended in 10 ml of precooled Buffer P1. After having added 10 ml of Buffer P2 the mixture was inverted 4-6 times and incubated for 5 min at RT before adding 10 ml of chilled Buffer P3. Thereafter the lysate was poured into a prepared QIAfilter Maxi Cartridge and incubated at RT for 10 min. During this time a QIAGEN-tip 500 was equilibrated by applying 10 ml of Buffer QBT and allowing the column to empty by gravity flow. The cell lysate was filtered into the QIAGEN-tip. The cleared lysate entered the resin by gravity flow and after washing with 2 x 30 ml Buffer QC the Plasmid DNA was eluted with 15 ml Buffer QF. After this the DNA was precipitated by adding 10.5 ml isopropanol and centrifuged at 4,000 rpm for 45 min at 4°C. The supernatant was discarded and the DNA pellet was washed with 5 ml ethanol (70%) and centrifuged at 4,000 rpm for 15 min. After air-drying the pellet the DNA was redissolved in H2O. <br> | 150 ml LB-Medium with 150 µl ampicillin was inoculated with 50 µl of bacteria culture which grew overnight on a shaker at 37°C. The plasmid DNA was isolated using QiAprep Spin MAxiprep kit from Qiagen and the protocol was followed. The overnight culture was centrifuged for 20 min at 4000 rpm at 4°C using an SLA 1500 Rotor. Afterwards the LB-medium was discarded and the pellet was homogeneously resuspended in 10 ml of precooled Buffer P1. After having added 10 ml of Buffer P2 the mixture was inverted 4-6 times and incubated for 5 min at RT before adding 10 ml of chilled Buffer P3. Thereafter the lysate was poured into a prepared QIAfilter Maxi Cartridge and incubated at RT for 10 min. During this time a QIAGEN-tip 500 was equilibrated by applying 10 ml of Buffer QBT and allowing the column to empty by gravity flow. The cell lysate was filtered into the QIAGEN-tip. The cleared lysate entered the resin by gravity flow and after washing with 2 x 30 ml Buffer QC the Plasmid DNA was eluted with 15 ml Buffer QF. After this the DNA was precipitated by adding 10.5 ml isopropanol and centrifuged at 4,000 rpm for 45 min at 4°C. The supernatant was discarded and the DNA pellet was washed with 5 ml ethanol (70%) and centrifuged at 4,000 rpm for 15 min. After air-drying the pellet the DNA was redissolved in H2O. <br> | ||
<br> | <br> | ||
| + | |||
| + | |||
| + | ===Plasmid-DNA isolation=== | ||
| + | 5 ml LB-Medium with 5 µl ampicillin was inoculated with single colonies which grew overnight on a shaker at 37°C. The plasmid DNA was isolated using QiAprep Spin Miniprep kit from Qiagen and following the manufacturer’s protocol. 4 ml of each overnight culture was pelleted in 2 ml microcentrifuge tubes during two steps of centrifugation at 13.000 rpm. Subsequently the pellet was resuspended in 250 µl of chilled buffer P1. 250 µl of lysis buffer P2 was added and the solution was mixed thoroughly by inverting the tube 4-6 times. After adding 350 µl of the neutralization Buffer N3 the solution was mixed immediately and thoroughly by inverting the tube 4-6 times. Thereafter the mixture was centrifuged. The supernatants were applied to a QIAprep column which was put in a 2 ml collection tube. It was centrifuged for 1 min at 13.000 rpm and the flow-through was discarded. After adding 500 µl of wash buffer PB, it was centrifuged for 1 min at 13.000 rpm and the flow-through was discarded. Once more, it was washed with 750 µl of wash buffer PE. In an additional centrifugation for 1 min at 13.000 rpm the residual wash buffer was removed. The QIAprep column was placed into a clean 1.5 ml microcentrifuge tube and the plasmid DNA was eluted in 30 µl ddH2O.<br> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | ===Purification of PCR product=== | ||
| + | One volume of buffer PBI was added to one volume of the PCR sample mix. The sample was applied to a QIAquick column which has been placed into a provided 2 ml collection tube. It was centrifuged for 1 min at 13.000 rpm and the flow-through was discarded and the column was placed in the same collection tube. After this 750 µl of buffer PE was added to wash the column. It was centrifuged for 1 min at 13.000 rpm. The flow-through was discarded and the column was placed in the same collection tube. It was centrifuged for 1 min at 13.000 rpm. Afterwards the QIAquick column was placed into a new 1.5 ml microcentrifuge tube and it was eluted with 40 µl ddH2O. | ||
| + | |||
| + | ===Sequencing=== | ||
| + | performed by the GATC Biotech company | ||
==Cell culture== | ==Cell culture== | ||
| Line 151: | Line 320: | ||
#* incubate 1 hour at 37°C | #* incubate 1 hour at 37°C | ||
#* wash five times | #* wash five times | ||
| - | + | #* add 100 µl substrate per well | |
| - | #* add | + | |
| - | + | ||
#* incubate at room temperature for 5 - 10 min | #* incubate at room temperature for 5 - 10 min | ||
| - | #* stop with | + | #* stop with 100µl 2M H<sub>2</sub>SO<sub>4</sub> per well |
| - | #* read at | + | #* read absorbance at 450 nm wavelength |
| + | |||
| + | |||
| + | ==== Buffers ==== | ||
| + | *'''Coating Buffer''' (0.1M NaHCO<sub>3</sub>, 0.1M Na<sub>2</sub>CO<sub>3</sub>, pH 9.5) | ||
| + | |||
| + | *'''Diluting/Blocking Buffer (db)''' (0.25 ml Tween 20, 30 g BSA, add 1x PBS to 500 ml) | ||
| + | |||
| + | *'''Wash Buffer''' (0.5 ml Tween 20, 1L PBS 1x) | ||
| + | |||
| + | |||
| + | :'''Calibration''' | ||
| + | * 960 µl db + 40 µl pooled plasma = 200 ng/ml | ||
| + | * add 500 µl of above into 500 µl of db = 100 ng/ml | ||
| + | * serial dilutions continued down to 1.5625 ng/ml | ||
| + | |||
| + | |||
| + | :'''Detection reagents''' | ||
| + | * TMB Substrate Kit (Pierce, Thermo Scientific) | ||
| + | * 2M H<sub>2</sub>SO<sub>4</sub> | ||
=== Microscopy === | === Microscopy === | ||
| Line 163: | Line 349: | ||
Single images were obtained using the Leica TCS SP5 confocal microscope and camera with the Leica AF6000 imaging software. GFP fluorescence was excited by Argon 488nm laser and measured at 520-560nm, BFP fluorescence was excited by UV laser at 405nm and measured at 440-460nm. Pictures were taken sequentially line by line in three different channels for GFP, BFP and bright field. | Single images were obtained using the Leica TCS SP5 confocal microscope and camera with the Leica AF6000 imaging software. GFP fluorescence was excited by Argon 488nm laser and measured at 520-560nm, BFP fluorescence was excited by UV laser at 405nm and measured at 440-460nm. Pictures were taken sequentially line by line in three different channels for GFP, BFP and bright field. | ||
| + | |||
| + | |||
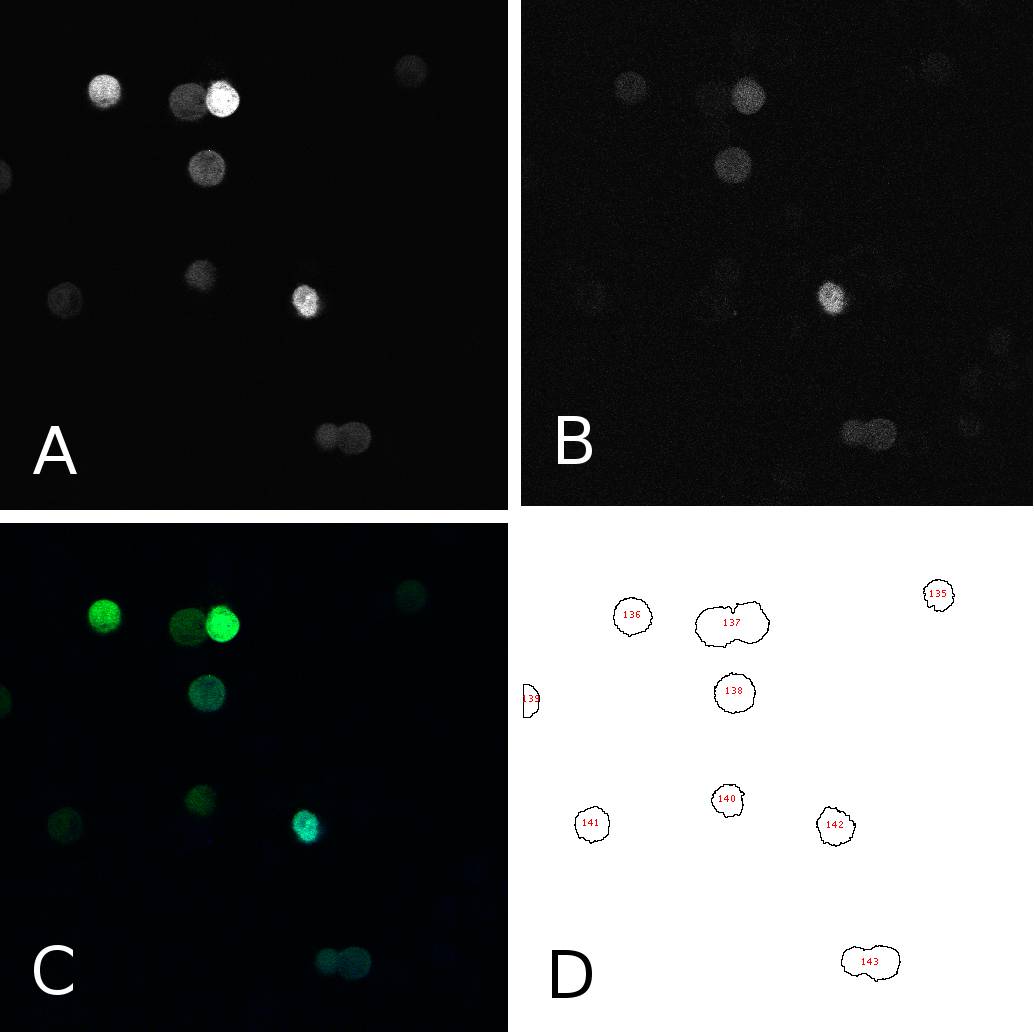
| + | [[Image:panel.jpg|thumb|600px|center|'''HeLa cells two days after transfection with miMeasure''' (A) fluorescence signal GFP channel, 8bit; (B) fluorescence signal BFP channel, 8bit; (C) merge of channels A and B, RGB (D) cells after segmentation and automated cell counting and annotation]] | ||
To analyze the fluorescence of single cells, we segmented the images using ImageJ. In 8bit pictures, we set the threshold for each channel to 50, thereby filtering the background. This allows us to annotate cells automatically using the “analyze particles” tool. We could now get the fluorescence intensity for each single cell on each channel (GFP or BFP) as an 8bit output, i.e. a value between 50 and 255. Panel 1 shows an example of one such image in different channels and after segmentation. From the data thus obtained, we calculated the GFP:BFP ratios for each cell using a simple algorithm. We could then visualize the mean of these rations in a bar plot or use all the data to calculate a linear regression curve. | To analyze the fluorescence of single cells, we segmented the images using ImageJ. In 8bit pictures, we set the threshold for each channel to 50, thereby filtering the background. This allows us to annotate cells automatically using the “analyze particles” tool. We could now get the fluorescence intensity for each single cell on each channel (GFP or BFP) as an 8bit output, i.e. a value between 50 and 255. Panel 1 shows an example of one such image in different channels and after segmentation. From the data thus obtained, we calculated the GFP:BFP ratios for each cell using a simple algorithm. We could then visualize the mean of these rations in a bar plot or use all the data to calculate a linear regression curve. | ||
| Line 180: | Line 369: | ||
We measured the knockdown of firefly luciferase using the Promega Dual Luciferase Reporter Assay. | We measured the knockdown of firefly luciferase using the Promega Dual Luciferase Reporter Assay. | ||
| - | The DLR™ Assay System provides an efficient mean of performing dual-reporter assays, where the activities of firefly (Photinus pyralis) and Renilla (Renilla reniformis) luciferases (RL) are measured sequentially from a single sample. Firefly and Renilla luciferases can be used as a good reporter system, as those two enzymes have dissimilar enzyme structures and substrate requirements. This allows for selective discrimination between their bioluminescent reactions. The firefly luciferase (FL) reporter is measured first by adding Luciferase Assay Reagent II (LAR II) to generate a stabilized luminescent signal. After quantifying the firefly luminescence, this reaction is quenched, and the Renilla luciferase reaction is simultaneously initiated by adding Stop & Glo® Reagent to the same tube. The Stop & Glo® Reagent also produces a stabilized signal from the Renilla luciferase, which decays slowly over the course of the measurement. Here, Renilla luciferase is used for normalization. The measurements were conducted on the Promega GLOMAX 96 Microplate Luminometer using the Promega standard protocol. | + | The DLR™ Assay System provides an efficient mean of performing dual-reporter assays, where the activities of firefly (<i>Photinus pyralis</i>) and Renilla (<i>Renilla reniformis</i>) luciferases (RL) are measured sequentially from a single sample. Firefly and Renilla luciferases can be used as a good reporter system, as those two enzymes have dissimilar enzyme structures and substrate requirements. This allows for selective discrimination between their bioluminescent reactions. The firefly luciferase (FL) reporter is measured first by adding Luciferase Assay Reagent II (LAR II) to generate a stabilized luminescent signal. After quantifying the firefly luminescence, this reaction is quenched, and the Renilla luciferase reaction is simultaneously initiated by adding Stop & Glo® Reagent to the same tube. The Stop & Glo® Reagent also produces a stabilized signal from the Renilla luciferase, which decays slowly over the course of the measurement. Here, Renilla luciferase is used for normalization. The measurements were conducted on the Promega GLOMAX 96 Microplate Luminometer using the Promega standard protocol ([https://2010.igem.org/Team:Heidelberg/Project/References#Materials_and_Methods Sherf et al., 1996]). <br> |
Twenty hours after transfection, cells were washed with 1x PBS and lysed using 1x Passive Lysis Buffer (5x stock solution diluted with distilled water), shaking for 30 minutes at 37°C. 10µl of the lysate were transferred to a white microplate (LumaPlate) as required for Luminometer measurements. | Twenty hours after transfection, cells were washed with 1x PBS and lysed using 1x Passive Lysis Buffer (5x stock solution diluted with distilled water), shaking for 30 minutes at 37°C. 10µl of the lysate were transferred to a white microplate (LumaPlate) as required for Luminometer measurements. | ||
| Line 189: | Line 378: | ||
The activity of the first luciferase (firefly) was measured by adding 25µl of LAR II reagent to the well. The enzyme reacts upon translation without further processing and oxidates beetle luciferin, resulting in photon emission that can be measured. In addition to beetle luciferin, the LAR II reagent contains coenzyme A, which accelerates the reaction and thus creates a prolonged luminescence signal. The luminescence was measured two seconds after addition of the reagent, for ten seconds. Afterwards, 25µl Stop & Glo reagent was added, which is able to quench the firefly luciferase activity and simultaneously contains the substrate for Renilla luciferase, coelenterazine. This second reaction also emits photons upon oxidation of the substrate. Addition of substrates and light emission measurements were conducted automatically by the GLOMAX Luminometer. | The activity of the first luciferase (firefly) was measured by adding 25µl of LAR II reagent to the well. The enzyme reacts upon translation without further processing and oxidates beetle luciferin, resulting in photon emission that can be measured. In addition to beetle luciferin, the LAR II reagent contains coenzyme A, which accelerates the reaction and thus creates a prolonged luminescence signal. The luminescence was measured two seconds after addition of the reagent, for ten seconds. Afterwards, 25µl Stop & Glo reagent was added, which is able to quench the firefly luciferase activity and simultaneously contains the substrate for Renilla luciferase, coelenterazine. This second reaction also emits photons upon oxidation of the substrate. Addition of substrates and light emission measurements were conducted automatically by the GLOMAX Luminometer. | ||
| - | |||
| - | |||
| - | |||
==== Consumables and Reagents ==== | ==== Consumables and Reagents ==== | ||
| - | LumaPlate, PerkinElmer, catalogue number 6005630 | + | LumaPlate, PerkinElmer, catalogue number 6005630<br> |
Promega Dual-Luciferase® Reporter Assay System, catalogue number E1910 | Promega Dual-Luciferase® Reporter Assay System, catalogue number E1910 | ||
| Line 201: | Line 387: | ||
Promega GLOMAX 96 Microplate Luminometer | Promega GLOMAX 96 Microplate Luminometer | ||
| - | === | + | === Plate reader measurements === |
Latest revision as of 02:14, 28 October 2010

|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"