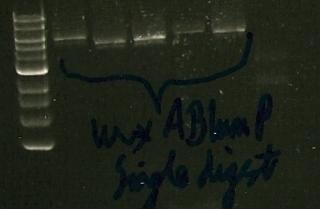Team:Edinburgh/Notebook/Blue light producer
From 2010.igem.org
(Difference between revisions)
Macbereska (Talk | contribs) (→Blue Light Producer) |
|||
| (12 intermediate revisions not shown) | |||
| Line 43: | Line 43: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Genomic">submitted parts</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Genomic">submitted parts</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Results#Genomic">results</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Genomic">results</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Project/Future">future | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Project/Future">the future</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Project/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Project/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 50: | Line 50: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial" class="dir">bacterial BRIDGEs</a> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial" class="dir">bacterial BRIDGEs</a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Core_repressilator">the | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Core_repressilator">the project</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_producer">red light</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_producer">red light</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_sensor">red sensor</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_sensor">red sensor</a></li> | ||
| Line 59: | Line 59: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Bacterial">submitted parts</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Bacterial">submitted parts</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Results#Bacterial">results</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Bacterial">results</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Future">future | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Future">the future</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 72: | Line 72: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Tools">tools</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Tools">tools</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Results#Modelling">results</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Modelling">results</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Future">future | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Future">the future</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 79: | Line 79: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human" class="dir">human BRIDGEs</a> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human" class="dir">human BRIDGEs</a> | ||
<ul> | <ul> | ||
| - | |||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/Communication">communication of science</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Communication">communication of science</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/ | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Branding">iGEM survey</a></li> |
| - | + | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/Conversations">conversations</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Conversations">conversations</a></li> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/Epic">the epic</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Epic">the epic</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/ | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/FutureApps">future applications</a></li> |
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Human | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Human">further thoughts</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 125: | Line 117: | ||
<div id="windowbox" style="border: .2em solid #660000; padding: 5px; position:fixed; top:50%; right:30px; width:8%;"> | <div id="windowbox" style="border: .2em solid #660000; padding: 5px; position:fixed; top:50%; right:30px; width:8%;"> | ||
| - | <span style="color:ivory;"> | + | <span style="color:ivory;">These <b>notes</b> correspond to the part of the <b>project</b> detailed <a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Blue_light_producer">here</a>.</span> |
</div> | </div> | ||
| Line 133: | Line 125: | ||
== Blue Light Producer == | == Blue Light Producer == | ||
<br> | <br> | ||
| + | '''5/7/2010''' | ||
| + | *LuxAB & LuxCDE operon agarose gel | ||
| + | * Lane1: About2.1.kbp | ||
| + | * Lane 2: The highest band- very faint- about 3.5kbp | ||
| + | [[Image:150.JPG]] | ||
'''13/7/2010''' | '''13/7/2010''' | ||
| Line 144: | Line 141: | ||
*The transformations of Edinbrick are fine but all the luxABs failed (grew blue, not white on Xgal and did not glow, as of 27/7)/ | *The transformations of Edinbrick are fine but all the luxABs failed (grew blue, not white on Xgal and did not glow, as of 27/7)/ | ||
*Still have some digested LuxAB-ediI so re-did ligation and transformation. | *Still have some digested LuxAB-ediI so re-did ligation and transformation. | ||
| - | [[Image: | + | [[Image:154.JPG]] |
{| border="1" | {| border="1" | ||
| Line 155: | Line 152: | ||
| LuxAB | | LuxAB | ||
|} | |} | ||
| - | + | [[Image:155.JPG]] | |
| - | + | ||
{| border="1" | {| border="1" | ||
| Lane 1 | | Lane 1 | ||
| Line 166: | Line 162: | ||
| LuxAB | | LuxAB | ||
|} | |} | ||
| + | |||
| + | '''29/7/2010''' | ||
| + | |||
| + | [[Image:156.jpg]] | ||
* The ligation had the same gel characteristics as previous luxAb-ediI ligations, e.x. it isn't visivle- not hopeful of this transformation working | * The ligation had the same gel characteristics as previous luxAb-ediI ligations, e.x. it isn't visivle- not hopeful of this transformation working | ||
| - | ''' | + | '''4/8/2010''' |
| + | * made minipreps of blue (negative colonies)- double digest and ran in comparison with pure edinbrickI | ||
| + | * LacZ band dissapears but colonies still grow blue (WTF) | ||
| + | * Has bands equaling to luxAB and EdiI but there was no glowing | ||
| + | |||
| + | [[Image:162.jpg]] | ||
'''12/8/2010''' | '''12/8/2010''' | ||
| Line 179: | Line 184: | ||
* Vector wa ssequenced and LumP is definately there. | * Vector wa ssequenced and LumP is definately there. | ||
| - | ''' | + | [[Image:160.jpg]] |
| + | |||
| + | Lanes 2-5: single digests of lumP | ||
| + | Lane 6: double digest of lumP | ||
| + | |||
| + | '''26/8/2010''' | ||
| + | |||
| + | * Marker (Lane 1), minipreps 6-10 luxABlumP (lanes 2-6) | ||
| + | [[Image:129.JPG]] | ||
| + | * single digests of LuxABlumP | ||
| + | [[Image:131.JPG]] | ||
| + | |||
| + | '''7/9/2010''' | ||
| + | *minipreps of luxCDE: samples 1&4 usable | ||
| + | |||
| + | {| border="1" | ||
| + | | Lane 1 | ||
| + | | Lane 2-6 | ||
| + | | Lanes 7 | ||
| + | |- | ||
| + | | Ladder | ||
| + | | luxCDE00:1-5 minipreps | ||
| + | | Ladder | ||
| + | |} | ||
| + | [[Image:140.JPG]] | ||
Latest revision as of 02:12, 28 October 2010
These notes correspond to the part of the project detailed here.
Blue Light Producer
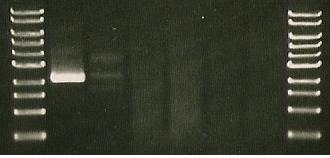
5/7/2010
- LuxAB & LuxCDE operon agarose gel
- Lane1: About2.1.kbp
- Lane 2: The highest band- very faint- about 3.5kbp
13/7/2010
Empty edinbrick vector for control.
- LuxAB + edinbrick ligation.
28/7/2010
an update on LuxAB:
- The transformations of Edinbrick are fine but all the luxABs failed (grew blue, not white on Xgal and did not glow, as of 27/7)/
- Still have some digested LuxAB-ediI so re-did ligation and transformation.
| Lane 1&15 | Lane 2 | Others |
| Ladder | EdiI | LuxAB |
| Lane 1 | Lane 2 +3 | Lanes 3-6 |
| Ladder | EdiI | LuxAB |
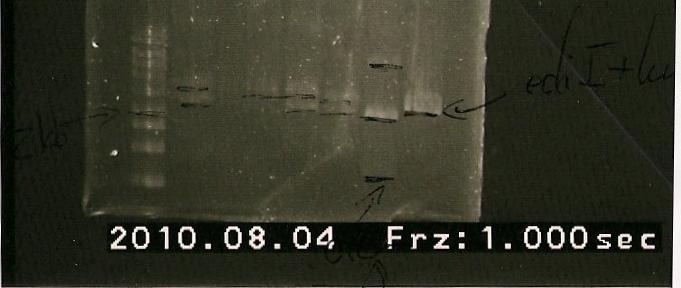
29/7/2010
- The ligation had the same gel characteristics as previous luxAb-ediI ligations, e.x. it isn't visivle- not hopeful of this transformation working
4/8/2010
- made minipreps of blue (negative colonies)- double digest and ran in comparison with pure edinbrickI
- LacZ band dissapears but colonies still grow blue (WTF)
- Has bands equaling to luxAB and EdiI but there was no glowing
12/8/2010 update on LumP:
- still getting pink/red colonies from transformants
- single digests give a band of about 3kb
- double digests give a band of about 2.5 kb -highly confusing
- and also, no sign on RFP despite red/pink growth.
- Vector wa ssequenced and LumP is definately there.
Lanes 2-5: single digests of lumP Lane 6: double digest of lumP
26/8/2010
- Marker (Lane 1), minipreps 6-10 luxABlumP (lanes 2-6)
- single digests of LuxABlumP
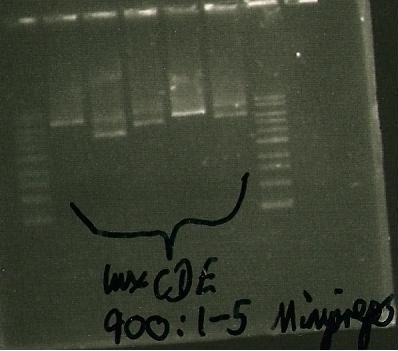
7/9/2010
- minipreps of luxCDE: samples 1&4 usable
| Lane 1 | Lane 2-6 | Lanes 7 |
| Ladder | luxCDE00:1-5 minipreps | Ladder |
 "
"