Team:Imperial College London/Modelling/Signalling/Detailed Description
From 2010.igem.org
(Difference between revisions)
(proof read) |
|||
| (One intermediate revision not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Imperial_College_London/Templates/Header}} | {{:Team:Imperial_College_London/Templates/Header}} | ||
| - | {{:Team:Imperial_College_London/Templates/ | + | {{:Team:Imperial_College_London/Templates/ModellingHeaderS}} |
{{:Team:Imperial_College_London/Templates/ModellingSignallingHeader}} | {{:Team:Imperial_College_London/Templates/ModellingSignallingHeader}} | ||
{| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| Line 73: | Line 73: | ||
|- | |- | ||
|In the above equations, the forwards reaction constants are represented by <html>k<sub>1,2,3,4,</sub> for the first, second, third and fourth reaction, respectively. The backwards reaction constants are represented by k<sub>-1,-2,-3,-4</sub></html>. | |In the above equations, the forwards reaction constants are represented by <html>k<sub>1,2,3,4,</sub> for the first, second, third and fourth reaction, respectively. The backwards reaction constants are represented by k<sub>-1,-2,-3,-4</sub></html>. | ||
| + | |- | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;" align="right"|[[Team:Imperial_College_London/Modelling/Signalling/Results_and_Conclusion | Click here for the results of this model...]] | ||
|} | |} | ||
Latest revision as of 23:00, 27 October 2010
| Modelling | Overview | Detection Model | Signaling Model | Fast Response Model | Interactions |
| A major part of the project consisted of modelling each module. This enabled us to decide which ideas we should implement. Look at the Fast Response page for a great example of how modelling has made a major impact on our design! | |
| Objectives | Description | Results | Constants | MATLAB Code |
| Detailed Description |
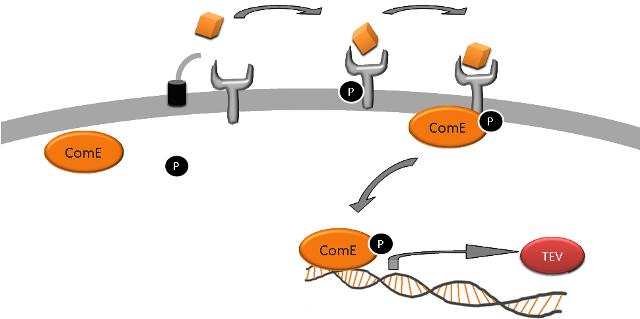
| We are assuming steady-state for AIP, Phosphate, ComD and ComE in our cell/in the solution. Hence, we can neglect production and degradation rates of ComD and ComE. |
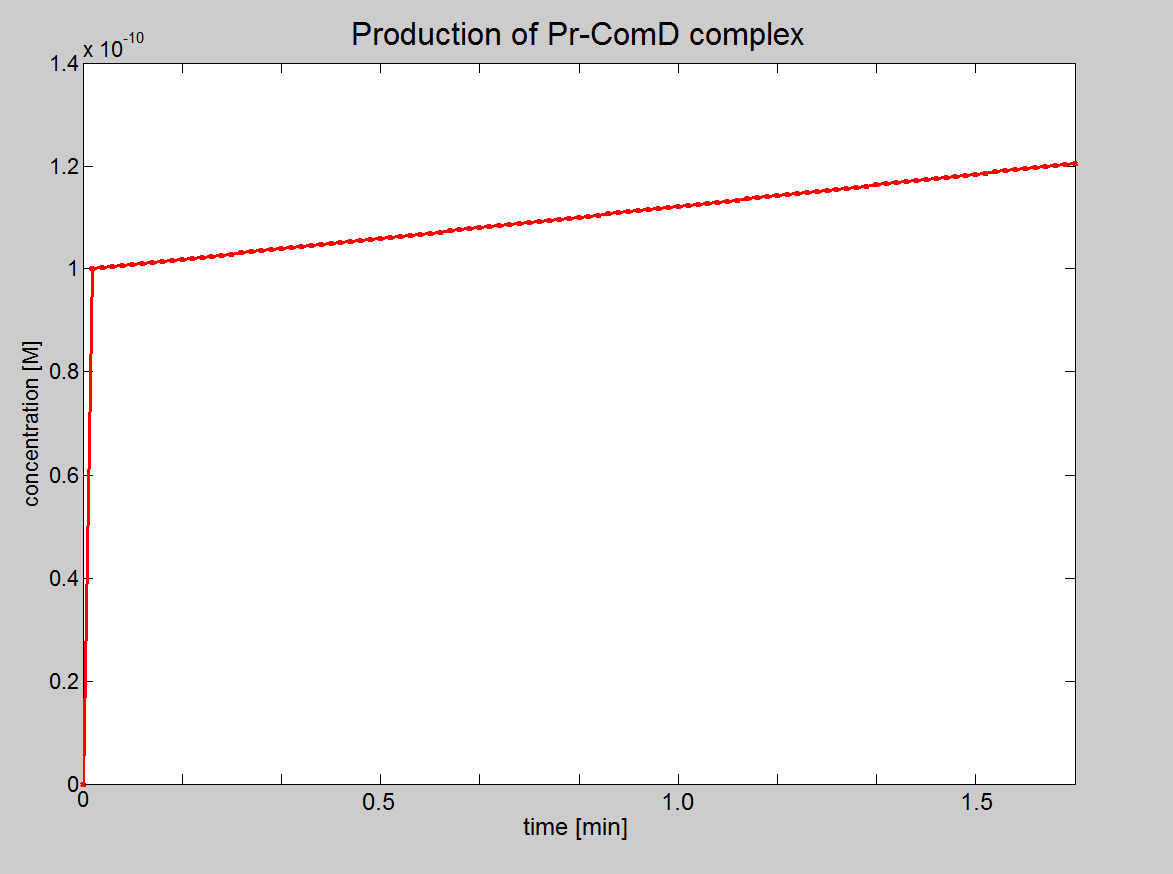
| Equation 1: AIP binds to ComD receptor to form a complex (AIP-ComD) |
| AIP + ComD ↔ AIP-ComD |
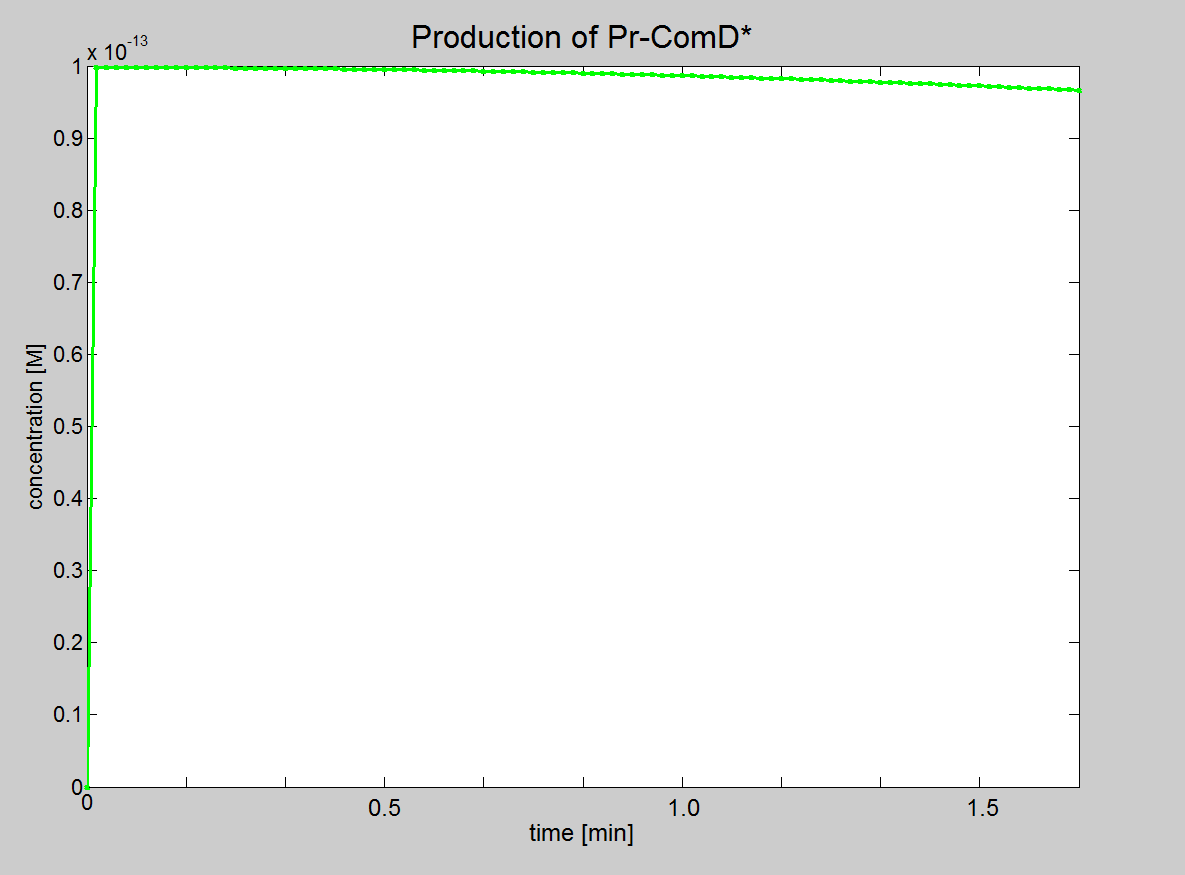
| Equation 2: Phosphate binds to the AIP-ComD complex to form another complex (AIP-ComD*) |
| AIP-ComD + Phosphate ↔ AIP-ComD* |
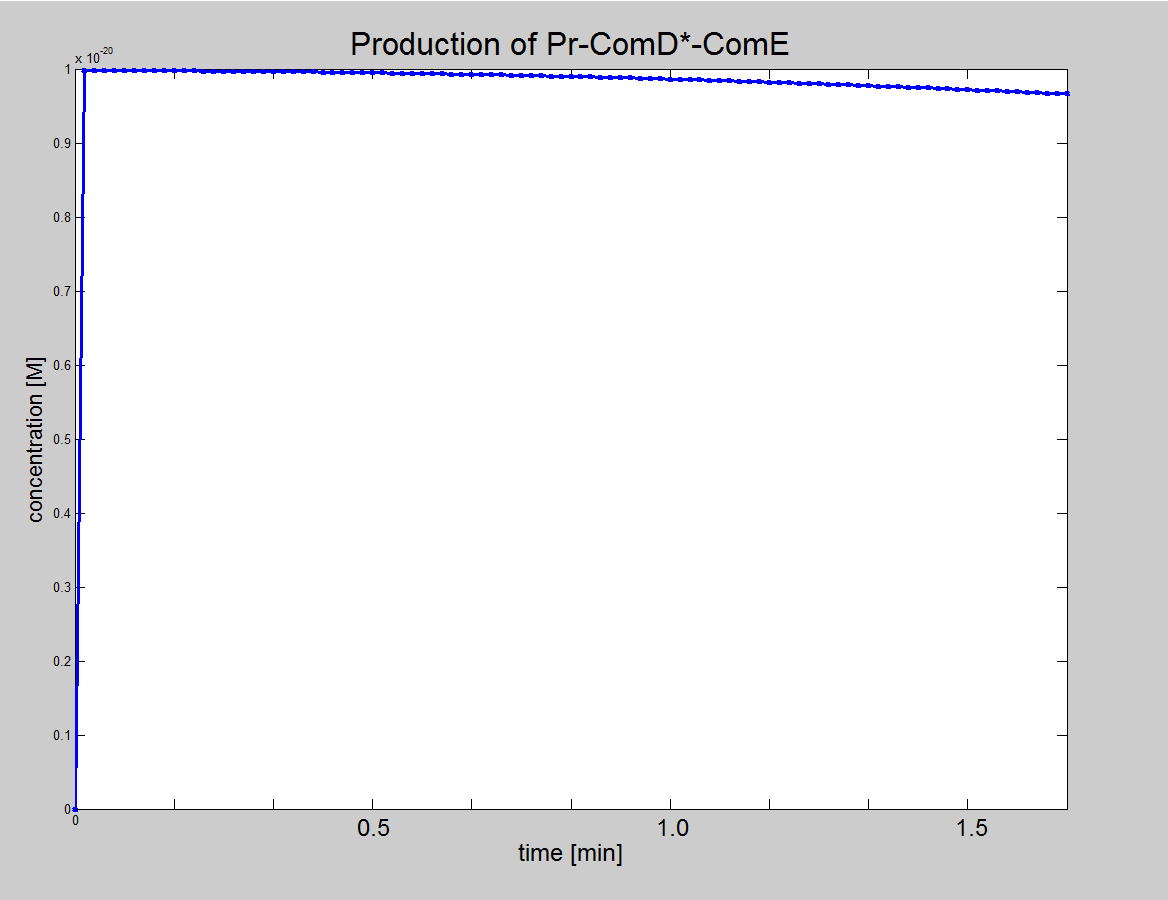
| Equation 3: ComE binds to the AIP-ComD* recepetor to form another complex (AIP-ComD*-ComE) |
| AIP-ComD* + ComE ↔ AIP-ComD*-ComE |
| Equation 4: Phosphate group on ComD binds to ComE and forms two products: phosphorylated ComE (ComE*) and AIP-ComD |
| AIP-ComD*-ComE ↔ AIP-ComD + ComE* |
| Using the Law of Mass Action, we can rewrite these 4 equations: |

|
| In the above equations, the forwards reaction constants are represented by k1,2,3,4, for the first, second, third and fourth reaction, respectively. The backwards reaction constants are represented by k-1,-2,-3,-4. |
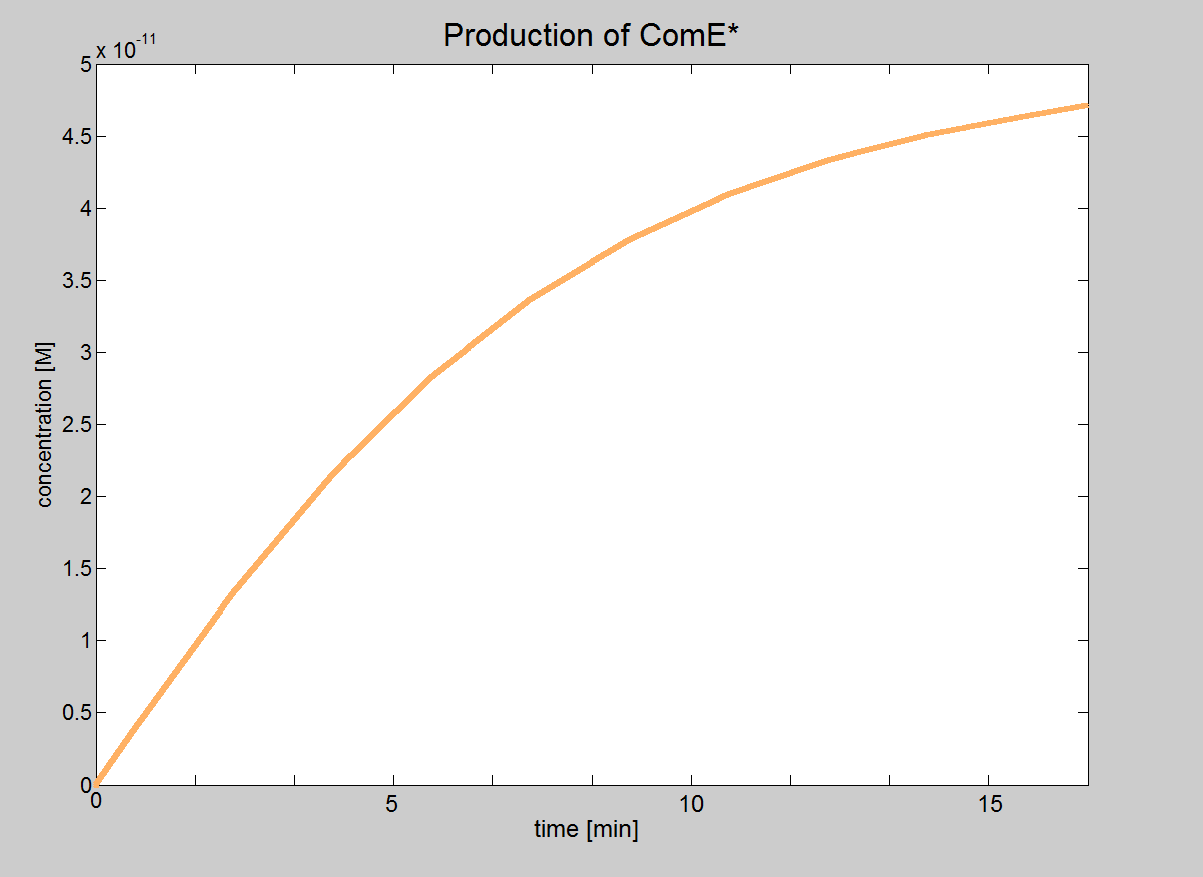
| Click here for the results of this model... |
 "
"