Team:Valencia/Parts
From 2010.igem.org
(→Transformation) |
(→Digestion) |
||
| Line 109: | Line 109: | ||
*1 μL PstI | *1 μL PstI | ||
*1 μL EcoRI | *1 μL EcoRI | ||
| - | *1,8 of insert | + | *1,8 of insert NMGR<sup>526</sup> (at 504,3 ng/ μL) |
*4 μL H buffer | *4 μL H buffer | ||
*32,2 μL water | *32,2 μL water | ||
| - | |||
===Ligation=== | ===Ligation=== | ||
Revision as of 22:02, 27 October 2010
Time goes by...
(El tiempo pasa...)
Follow us:

Our main sponsors:

Our institutions:

Visitor location:
Submitted Biobricks
Parts
We placed the first prion-based inheritance element construction into a standard biological parts, also called BioBricks. These are nucleic acid coding molecular biological functions, along with the associated information defining and describing the parts. You can find more information about this on the website of [http://biobricks.org/ The BioBricks Foundation].
Using these molecules any synthetic biologist or biological engineer can program living organisms. All our parts are available to anyone for free via MIT's [http://parts.mit.edu Registry of Standard Biological Parts].
We also placed the PM2 gene, coding the LEA3 protein, in another Biobrick, and probed that it conferes to our E. coli protection against extreme temperature.
<groupparts>iGEM010 Valencia</groupparts>
LEA
Function
PM2 corresponds to the LEA3 protein and it comes from soybean. LEA proteins are known for their roles in plant embryogenesis, as important dessication-resistance factors. It has also been shown that they confer tolerance under several stress conditions in Escherichia coli. You can read more about it in our wiki and in several references, as, for example, the following: Liu et al. (2010) Curr Microbiol 60: 373-378. PM2 can be useful as a general stress-resistance factor. It could improve survival under low or high temperatures, water stress and maybe other atypical conditions.
Original source
Our PM2 was inserted into the pET28a vector and transformed into E. coli BL21 Star cells. We have to thank professor Yizhi Zheng and Yun Liu (College of Life Science, Shenzhen University. Guangdong, China) for sending us the plasmid with the gene, and, in consequence, let us develop our project.
Sequence
The sequence of PM2 is the following one:
Then, in order to amplificate it and clone it into the pSB1C3, we used the following primer:
- Forward actagtagcggccgctgcagATGGCGTCCAAGAAAC
- Reverse tctagaagcggccgcgaattcTGCGTCTATATATAC
(capital letters indicate the region of the sequence that pairs with the coding sequence of PM2).
Prionic Switch
Components
The switch is formed by two different parts: the activator and the reporter. The activator part is a construction of two fragments: the NM domains of the protein Sup35, which confers to the protein the prionic activity, and the GR526 portion, which contains the DNA-binding and transcription-activation domains. The ligand-binding domain of the protein GR was eliminated, decoupling the response of the protein to the presence of glucocorticoids, and thus generating a constitutive transcription activation factor. The normal activity of this protein results in the activation of the genes preceded by the GRE (Glucocorticoid Response Element). When exposed to heat shock or other stress conditions, the NM domains start the prionic activity, eventually inhibiting the activation of transcription.
This part was amplified by using the primers indicated by Li and Lindquist (2000), together with the sequence recommended to use for the ligation protocol with the plasmid pSB1C3. Those primers are:
- Forward actagtagcggccgctgcagATGTCGGATTCAAAC
- Reverse tctagaagcggccgcgaattcTCCTGCAGTGGCTTG
(again, capital letters represent the region that pairs with the coding sequence of NMGR526).
The second part consists of the GRE followed by the reporter gene. In our experiments, we used LacZ for this purpose. The amplification of this part could not be made because of some problems found when trying to find the sequence of the GRE.
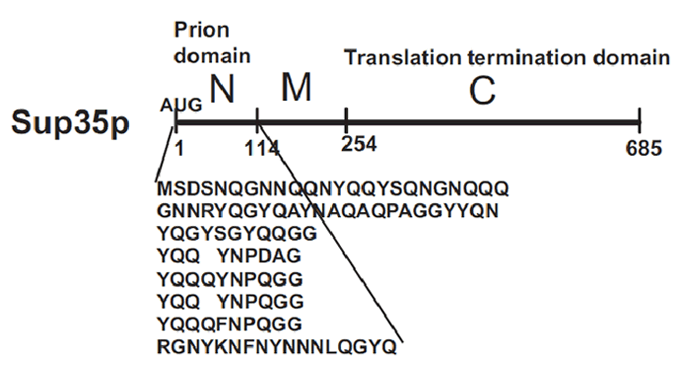
Sup35p
[PSI+] is a non-Mendelian trait of Saccharomyces cerevisae that supress nonsense codons. This phenotype is due to a self-replication conformation (prion state) of a protein encoded by the gene Sup35. This protein, Sup35p, is the yeast eukariotyc release factor 3 (eRF3) and forms the translation termination complex with Sup45p (eRF1). The function of Sup45p is releasing the nascent polypeptide chain from the ribosome through GTP hydrolysis when Sup45p recognize a stop codon.
Sup35p is 685 amino acids long and has three distinct parts (Fig.2). The NH2-terminus (N) is termed the prion-forming domain (PrD) because plays a critical role in Sup35p’s changes in proteic conformation and it is responsible for its prion behaviour. This domain is 114 amino acids long and has a high content in glutamine and asparagine. The middle region (M) provides a solubilizing and/or spacing function. Finally, the COOH-terminus (C) is responsible for the translation-termination activity.
In [PSI+] cells, most Sup35p is insoluble and nonfunctional, causing an increase in the translational read-through of stop codons. This trait is heritable because Sup35p in the amyloid state as every prion influences new Sup35p to adopt the same conformation and passes from mother cell to daughter. In [psi-] cells, the translation-termination factor Sup35p is soluble and functional.
Behaviour
Sup35p is a subunit of the translation termination complex. Its prionic nature has been proposed to have some effect on the stress response, as a possible mechanism to obtain modified genetic expression products. When the prionic conformation is activated, the termination of translation is less effective and thus new longer proteins form (True and Lindquist, 2000, Nature, 407: 477-483; Tyedmers et al., 2008, PLoS Biology, 6: e294). When the sequence corresponding to the NM domains of Sup35p is fused to other gene, the protein resulting of this construction acquires the prionic behaviour (Li and Lindquist, 2000, Science, 287: 661-664).
On the other side, GR (Glucocorticoid Receptor) activates the transcription of genes preceded by GRE (Glucocorticoid Receptor Element) when steroid hormones are present (Heitzer et al., 2007, Rev. Endocr. Metab. Disord., 8: 321-330). However, it becomes a constitutive transcription activator when it lacks its C terminal ligand-binding domain (Schena and Yamamoto, 1988, Science, 241: 965-967). Because of the length of amino acids of the cut protein, this short version of GR is named GR526.
Li and Lindquist (2000) showed that the fused protein (NMGR526) is a functional constitutive transcription activator. In addition, when the prionic conformation is reached because of the presence of a certain stimulus, NMGR526 is no longer capable of inducing the activation of the gene preceded by GRE. Tyedmers et al. (2000) checked the conditions that trigger the prionic conformation and they found that heat shock is a significantly relevant factor. The cells in which the prionic conformation is induced, the process is promoted in an autocatalytic manner and all the protein is found in the prionic conformation. The cells resulting show the phenotype [PSI+].
It is important to note that the rate of the change of conformation is not equal to zero even at optimal growth conditions, and that not all the cells become [PSI+]. The rate of spontaneous activation of the switch is thought to be around 10-6 or 10-7 (Alberti et al., 2009, Cell, 137: 146-158). This process is promoted under heat stress (Tyedmers et al., 2008), probably because of the important role of heat shock proteins like Hsp104 in the formation and maintenance of the amyloid fiber (Halfmann et al., 2009, Trends in Cell Biology). These approximate rates will have very important implications for the yeastworld model that we briefly describe in the following subsection, and with more detail in the Modeling section.
Original source
We have to thank D. Susan Lindquist (Whitehead Institute for Biomedical Research / Howard Hughes Medical Institute / Department of Biology, MIT) for sending us the prionic system transformed E.coli strains.
Making our Biobricks
After a lot of attempts trying to make our BioBricks (we had some dificulties at different steps) we achieved our purpose: we built 2 BioBricks and we sent them to de Registry.
PCR
First of all, we needed enough amounts of DNA in order to insert them as our Biobrick into the pSB1C3 provided by the Registry. We had purified DNA from PM2 E.coli strains and from NMGR526 E.coli strains. Using these as our DNA template we amplified the quoted genes: PM2 and NMGR526. The chosen program was:
- A first denaturation cycle: 94ºC 3min
- 30 amplification cycles, made up of:
- 94ºC 30s
- 55ºC 1min
- 72ºC 1min
- Final step: 72ºC 7min
Digestion
Next step was the digestion of the amplified material with the enzimes EcoRI and PstI, keeping the reaction tubes during 4-5 hours at 37ºC with:
PM2
- 1 μL PstI
- 1μL EcoRI
- 2,5 μL of insert PM2 (at 370,9 ng/ μL)
- 4 μL H buffer
- 31,5 μL water
NMGR526
- 1 μL PstI
- 1 μL EcoRI
- 1,8 of insert NMGR526 (at 504,3 ng/ μL)
- 4 μL H buffer
- 32,2 μL water
Ligation
After purificating the DNA using a simple etanol precipitation protocol, we resuspended the DNA into 12 μL of water. Then, we put both plasmid and PM2 in one tube, and both plasmid and NMGR526 in another one, using these amounts of components:
- 1 μL T4 ligase
- 3 μL pSB1C3
- 4 μL ligation buffer
- Whole 12 μL of our DNA
We kept it O/N in the fridge at 4ºC.
Transformation
We transformed DH5α E.coli competent strain with our ligation product using a standard heat shock transformation protocol.
Miniprep
Before the tranformation ending O/N step, we noticed that we had grown colonies in cloramphenicol LB plaques, so we used them to make a plasmidic DNA extraction (Miniprep: High Pure Plasmid Isolation Kit, Roche. 11 754 785 001). In order to check the process had been successful, we made an electrophoresis gel, putting four careers:
- Ligation product digested with EcoRI
- Ligation product digested with PstI
- Ligation product digested with EcoRI and PstI
- Ligation product without any restriction enzyme
 "
"