Team:WITS-South Africa/The Machine
From 2010.igem.org
| (8 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{{Template:WITS-South_Africa_Main_Menu_0910}} | {{Template:WITS-South_Africa_Main_Menu_0910}} | ||
| + | <div style="padding:40px;"> | ||
| + | |||
| + | =Project Overview= | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Our machine was designed as a proof of concept that commensal bacteria, which live normally and harmlessly on the human body, can be engineered to function as diagnostic and prophylactic devices against pathogens which are otherwise difficult to detect or control. Basically we wish to explore the possibility that commensal bacteria can be used as a biological “suit of armour” against disease. | Our machine was designed as a proof of concept that commensal bacteria, which live normally and harmlessly on the human body, can be engineered to function as diagnostic and prophylactic devices against pathogens which are otherwise difficult to detect or control. Basically we wish to explore the possibility that commensal bacteria can be used as a biological “suit of armour” against disease. | ||
| - | We decided to focus on Human Papillomavirus, which is a sexually transmitted infection which causes cervical cancer. HPV invades the vaginal mucosa, which is where commensal ''Lactobacillus gasseri'' and various other Lactic Acid Bacteria can be found in high numbers in healthy females, making ''L. gasseri'' a good candidate as a chassis for our protective machine. | + | We decided to focus on Human Papillomavirus, which is a sexually transmitted infection which causes cervical cancer. HPV invades the vaginal mucosa, which is where commensal ''Lactobacillus gasseri'' and various other Lactic Acid Bacteria can be found in high numbers in healthy females, making ''L. gasseri'' a good candidate as a chassis for our protective machine. Although we acquired a strain of ''L.gasseri'', we were unable to test the machine in it due to a lack of appropriate shuttle vectors for cloning purposes. Thus, we elected to use ''Bacillus subtilis'' as a model Gram-positive organism, for initial proof of concept. |
| + | |||
| + | == '''Machine Schematic''' == | ||
| + | |||
| + | |||
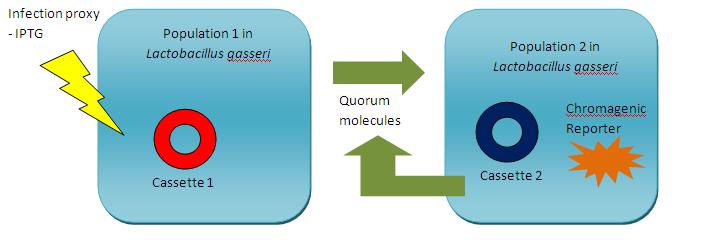
| + | This schematic diagram illustrates how our machines would function in tandem to detect and report the presence of a viral infection. The commensal bacterial chassis would contain either of two machines and would need to exist as a mixed population. One machine, would detect the presence of the viral infection and use quorum sensing peptides to signal to the other machine that a virus has been spotted. This machine would then produce a coloured reporter which would be detectable by the host. | ||
| + | |||
| + | |||
| + | [[Image:Schematic_wiki.JPG]] | ||
| + | |||
| + | |||
| + | |||
| + | ==Desired behaviour of our machines== | ||
When designing the machine we considered the following: | When designing the machine we considered the following: | ||
| Line 23: | Line 33: | ||
| - | '''The nature of the response – How will the machine alert an individual that she has been exposed to the virus?''' | + | '''The nature of the response – How will the machine alert an individual that she has been exposed to the virus?''' |
Traditional whole cell biosensors have been engineered to respond to one or more stimuli with the expression of fluorescent proteins such as firefly luciferase or GFP. When these biosensors are added to an environmental sample, such as potentially contaminated water, and then the sample is viewed under a fluorescent microscope, a positive response can easily be detected. However, if the biosensor is part of a population of commensal bacteria in the vaginal mucosa, this would not be appropriate or useful! Thus, we wanted our machine to respond by producing an output easily visible to the naked eye. Preferably, an individual could monitor the response by herself, with no need to go to a doctor for examination or undergo a complicated procedure. Thus, we decided to experiment with Cambridge’s winning iGEM 09 project – E.chromi biobricks which cause bacteria to produce easily visible chromogenic reporter molecules. Simultaneous production of a reporter molecule and one which blocks, contains or neutralises the viral particle would be ideal, but for the purposes of this project, we decided to focus on the reporting mechanism, with the option to add in a neutralising molecule at a later stage. | Traditional whole cell biosensors have been engineered to respond to one or more stimuli with the expression of fluorescent proteins such as firefly luciferase or GFP. When these biosensors are added to an environmental sample, such as potentially contaminated water, and then the sample is viewed under a fluorescent microscope, a positive response can easily be detected. However, if the biosensor is part of a population of commensal bacteria in the vaginal mucosa, this would not be appropriate or useful! Thus, we wanted our machine to respond by producing an output easily visible to the naked eye. Preferably, an individual could monitor the response by herself, with no need to go to a doctor for examination or undergo a complicated procedure. Thus, we decided to experiment with Cambridge’s winning iGEM 09 project – E.chromi biobricks which cause bacteria to produce easily visible chromogenic reporter molecules. Simultaneous production of a reporter molecule and one which blocks, contains or neutralises the viral particle would be ideal, but for the purposes of this project, we decided to focus on the reporting mechanism, with the option to add in a neutralising molecule at a later stage. | ||
| - | '''The strength of the response needed for detection''' | + | '''The strength of the response needed for detection''' |
| + | [[Image:DSC02619a.JPG|300px|right]] | ||
If a viral particle is present in the vaginal mucosa, and one, or even a few local bacterial biosensors detects it, the colour change in those cells will not be sufficient for detection by the host. In order for our machine to be of practical use, infection needs to trigger a coordinated response from the entire population, switching every biosensor on. For this we explored the use of quorum sensing molecules, which are a method of bacterial communication which can directly activate gene expression. Gram-positive bacteria, such as ''L. gasseri'', produce species specific quorum peptides which can be used to create a positive feedback loop to propagate an initial infection signal amongst an entire bacterial population. | If a viral particle is present in the vaginal mucosa, and one, or even a few local bacterial biosensors detects it, the colour change in those cells will not be sufficient for detection by the host. In order for our machine to be of practical use, infection needs to trigger a coordinated response from the entire population, switching every biosensor on. For this we explored the use of quorum sensing molecules, which are a method of bacterial communication which can directly activate gene expression. Gram-positive bacteria, such as ''L. gasseri'', produce species specific quorum peptides which can be used to create a positive feedback loop to propagate an initial infection signal amongst an entire bacterial population. | ||
| - | + | ||
| - | '''How to switch the machine off again after infection has been reported''' | + | '''How to switch the machine off again after infection has been reported''' |
A continuous “on” state is obviously not desirable, given that the eventual effect of machine activation would be an entire bacterial population in the vaginal mucosa that is producing a bright, visible colour. Once infection has been noted by the host, allowing them to seek medical treatment, the population needs to return to an off, non visible state – Firstly to avoid discomfort or embarrassment for the host and secondly to ensure that the machine can be used to further detect any subsequent infections. A repressor protein, which can switch off expression of the quorum molecule and shut down the positive-feedback loop, creating a negative-feedback loop, was selected. This would stop expression of the chromogenic reporter, and eventually, the bacterial cells would return to their normal colour. | A continuous “on” state is obviously not desirable, given that the eventual effect of machine activation would be an entire bacterial population in the vaginal mucosa that is producing a bright, visible colour. Once infection has been noted by the host, allowing them to seek medical treatment, the population needs to return to an off, non visible state – Firstly to avoid discomfort or embarrassment for the host and secondly to ensure that the machine can be used to further detect any subsequent infections. A repressor protein, which can switch off expression of the quorum molecule and shut down the positive-feedback loop, creating a negative-feedback loop, was selected. This would stop expression of the chromogenic reporter, and eventually, the bacterial cells would return to their normal colour. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
Latest revision as of 21:12, 27 October 2010

Project Overview
Our machine was designed as a proof of concept that commensal bacteria, which live normally and harmlessly on the human body, can be engineered to function as diagnostic and prophylactic devices against pathogens which are otherwise difficult to detect or control. Basically we wish to explore the possibility that commensal bacteria can be used as a biological “suit of armour” against disease.
We decided to focus on Human Papillomavirus, which is a sexually transmitted infection which causes cervical cancer. HPV invades the vaginal mucosa, which is where commensal Lactobacillus gasseri and various other Lactic Acid Bacteria can be found in high numbers in healthy females, making L. gasseri a good candidate as a chassis for our protective machine. Although we acquired a strain of L.gasseri, we were unable to test the machine in it due to a lack of appropriate shuttle vectors for cloning purposes. Thus, we elected to use Bacillus subtilis as a model Gram-positive organism, for initial proof of concept.
Machine Schematic
This schematic diagram illustrates how our machines would function in tandem to detect and report the presence of a viral infection. The commensal bacterial chassis would contain either of two machines and would need to exist as a mixed population. One machine, would detect the presence of the viral infection and use quorum sensing peptides to signal to the other machine that a virus has been spotted. This machine would then produce a coloured reporter which would be detectable by the host.
Desired behaviour of our machines
When designing the machine we considered the following:
The ability of the bacteria to detect and respond to a virus/viral element
Engineering a bacterium to alter its gene expression based on recognition of a specific virus is a major difficulty. For the purposes of this project we chose to focus, instead, on building and tweaking the ability of the machine to respond in certain ways to a specific (but non-viral) stimulus as a “proxy” for infection. Once this was achieved, it would be possible to investigate how to make the machine specific for HPV.
The nature of the response – How will the machine alert an individual that she has been exposed to the virus?
Traditional whole cell biosensors have been engineered to respond to one or more stimuli with the expression of fluorescent proteins such as firefly luciferase or GFP. When these biosensors are added to an environmental sample, such as potentially contaminated water, and then the sample is viewed under a fluorescent microscope, a positive response can easily be detected. However, if the biosensor is part of a population of commensal bacteria in the vaginal mucosa, this would not be appropriate or useful! Thus, we wanted our machine to respond by producing an output easily visible to the naked eye. Preferably, an individual could monitor the response by herself, with no need to go to a doctor for examination or undergo a complicated procedure. Thus, we decided to experiment with Cambridge’s winning iGEM 09 project – E.chromi biobricks which cause bacteria to produce easily visible chromogenic reporter molecules. Simultaneous production of a reporter molecule and one which blocks, contains or neutralises the viral particle would be ideal, but for the purposes of this project, we decided to focus on the reporting mechanism, with the option to add in a neutralising molecule at a later stage.
The strength of the response needed for detection
If a viral particle is present in the vaginal mucosa, and one, or even a few local bacterial biosensors detects it, the colour change in those cells will not be sufficient for detection by the host. In order for our machine to be of practical use, infection needs to trigger a coordinated response from the entire population, switching every biosensor on. For this we explored the use of quorum sensing molecules, which are a method of bacterial communication which can directly activate gene expression. Gram-positive bacteria, such as L. gasseri, produce species specific quorum peptides which can be used to create a positive feedback loop to propagate an initial infection signal amongst an entire bacterial population.
How to switch the machine off again after infection has been reported
A continuous “on” state is obviously not desirable, given that the eventual effect of machine activation would be an entire bacterial population in the vaginal mucosa that is producing a bright, visible colour. Once infection has been noted by the host, allowing them to seek medical treatment, the population needs to return to an off, non visible state – Firstly to avoid discomfort or embarrassment for the host and secondly to ensure that the machine can be used to further detect any subsequent infections. A repressor protein, which can switch off expression of the quorum molecule and shut down the positive-feedback loop, creating a negative-feedback loop, was selected. This would stop expression of the chromogenic reporter, and eventually, the bacterial cells would return to their normal colour.
 "
"