Team:VT-ENSIMAG/Genothreat
From 2010.igem.org
| (13 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
=Global presentation= | =Global presentation= | ||
[[Image:Compdna.png|right|200px]] | [[Image:Compdna.png|right|200px]] | ||
| - | The software, GenoTHREAT takes a DNA sequence on input, and in return told the user if it's a dangerous sequence which need further investigation or if the sequence can be | + | The software, GenoTHREAT takes a DNA sequence on input, and in return told the user if it's a dangerous sequence which need further investigation or if the sequence can be synthesized. |
| - | It follows the algorithm given by the federal guidelines, and so uses the Blast software for sequence | + | It follows the algorithm given by the federal guidelines, and so uses the Blast software for sequence alignment. |
For managing the sequence (six-frame translation, reversed frame, extracting sub-sequence), we use Biojava, a bio-informatics toolbox ([http://www.biojava.org/wiki/Main_Page 1]). | For managing the sequence (six-frame translation, reversed frame, extracting sub-sequence), we use Biojava, a bio-informatics toolbox ([http://www.biojava.org/wiki/Main_Page 1]). | ||
GenoTHREAT has been coded in Java, and is working on both Windows and Linux distribution. | GenoTHREAT has been coded in Java, and is working on both Windows and Linux distribution. | ||
| - | We have | + | We have developed different versions in order to optimize the time execution or the CPUs' utilization (Central Processing Units) according to the use ([[#Different implementations]]). |
=General algorithm= | =General algorithm= | ||
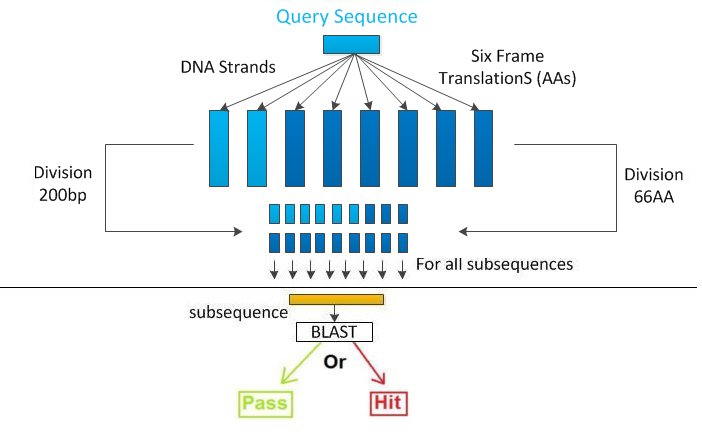
| - | Following the federal guideline, the first step is to do the six frame translation ([[Team:VT_Ensimag_2010-Biosecurity/sixframe|See More]]) in order to have the corresponding amino-acid strands. We also keep the initial nucleotid strand and its reversed DNA sequence ([[/Why?]]). So we have now 8 sequences to screen (6 amino-acid, 2 nucleotides). | + | Following the federal guideline, the first step is to do the six frame translation ([[Team:VT_Ensimag_2010-Biosecurity/sixframe|See More]]) in order to have the corresponding amino-acid strands. We also keep the initial nucleotid strand and its reversed DNA sequence ([[Team:VT_Ensimag_2010_Biosecurity/DNAkeep|Why?]]). So we have now 8 sequences to screen (6 amino-acid, 2 nucleotides). |
Then, we divide each sequence in 200bps nucleotide or 66 amino-acid subsequences. | Then, we divide each sequence in 200bps nucleotide or 66 amino-acid subsequences. | ||
<br> | <br> | ||
Each sub-sequence is then blasted, and we extract the relevant information from the blast output. | Each sub-sequence is then blasted, and we extract the relevant information from the blast output. | ||
For each sub-sequence, we have to decide if it's a hit or not ([[Team:VT_Ensimag_2010-Biosecurity/BestMatch|See More]]). | For each sub-sequence, we have to decide if it's a hit or not ([[Team:VT_Ensimag_2010-Biosecurity/BestMatch|See More]]). | ||
| - | [[Image:VTENSI_Algo. | + | [[Image:VTENSI_Algo.jpeg|center|400px]] |
| - | If one sub-sequence is a hit for our algorithm, then it means that a 200bps sequence potentially dangerous has been detected. The sequence will be so flagged, and a result file containing all the select agent found and some more information will be created. | + | If one sub-sequence is a hit for our algorithm, then it means that a 200bps sequence potentially dangerous has been detected. The sequence will be so flagged, and a result file containing all the select agent found and some more information will be created. If any of the sub-sequences is considered as a sequence of concern, then the sequence is accepted and can be synthesized. |
| + | |||
| + | <br> | ||
| + | [[Image:VTENSI_Algogen.jpg|frame|center|<br> ''Global algorithm of GenoTHREAT'']] | ||
=Different options= | =Different options= | ||
| Line 23: | Line 26: | ||
=Different implementations= | =Different implementations= | ||
| - | Our main issue in implementing the software is the | + | Our main issue in implementing the software is the resources-consuming(time and computational resources) of blast. In order to get rid of it, we have tried different implementation of our algorithm. See the description and comparison in [[Team:VT_Ensimag_2010-Biosecurity/Implementations|Different implementations]]. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | =Availability= | ||
| + | Due to GenoTHREAT sensible aspect on security, we are unfortunately not allowed to make it publicly available. You can nonetheless contact us and ask for it. You will need to identify yourself so that we deliver it to you. We apologise for this inconvenience. | ||
}} | }} | ||
Latest revision as of 19:42, 27 October 2010
GenoTHREAT
|
Global presentationThe software, GenoTHREAT takes a DNA sequence on input, and in return told the user if it's a dangerous sequence which need further investigation or if the sequence can be synthesized. It follows the algorithm given by the federal guidelines, and so uses the Blast software for sequence alignment. For managing the sequence (six-frame translation, reversed frame, extracting sub-sequence), we use Biojava, a bio-informatics toolbox ([http://www.biojava.org/wiki/Main_Page 1]). GenoTHREAT has been coded in Java, and is working on both Windows and Linux distribution. We have developed different versions in order to optimize the time execution or the CPUs' utilization (Central Processing Units) according to the use (#Different implementations). General algorithmFollowing the federal guideline, the first step is to do the six frame translation (See More) in order to have the corresponding amino-acid strands. We also keep the initial nucleotid strand and its reversed DNA sequence (Why?). So we have now 8 sequences to screen (6 amino-acid, 2 nucleotides).
Then, we divide each sequence in 200bps nucleotide or 66 amino-acid subsequences.
If one sub-sequence is a hit for our algorithm, then it means that a 200bps sequence potentially dangerous has been detected. The sequence will be so flagged, and a result file containing all the select agent found and some more information will be created. If any of the sub-sequences is considered as a sequence of concern, then the sequence is accepted and can be synthesized.
Different optionsAround this algorithm, we have developed different options to use the sequence screening. We can use it to just screen one sequence, or a group of sequence from our test database, or screen sequence from the iGEM registry or GenoCAD database. The parameter on some versions can also be changed in creating a new set of parameters. Different implementationsOur main issue in implementing the software is the resources-consuming(time and computational resources) of blast. In order to get rid of it, we have tried different implementation of our algorithm. See the description and comparison in Different implementations. AvailabilityDue to GenoTHREAT sensible aspect on security, we are unfortunately not allowed to make it publicly available. You can nonetheless contact us and ask for it. You will need to identify yourself so that we deliver it to you. We apologise for this inconvenience. |
 "
"