Team:BCCS-Bristol/Wetlab/K381001 Construction
From 2010.igem.org
m |
|||
| (10 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{:Team:BCCS-Bristol/Header}} | |
| - | + | =Constructing Our Biobrick - K381001= | |
| - | + | ||
| - | Construction of the | + | ==Overview== |
| - | * Generating larger quantities of both our component | + | |
| - | * Digesting each of these | + | {{:Team:BCCS-Bristol/newtoc}} |
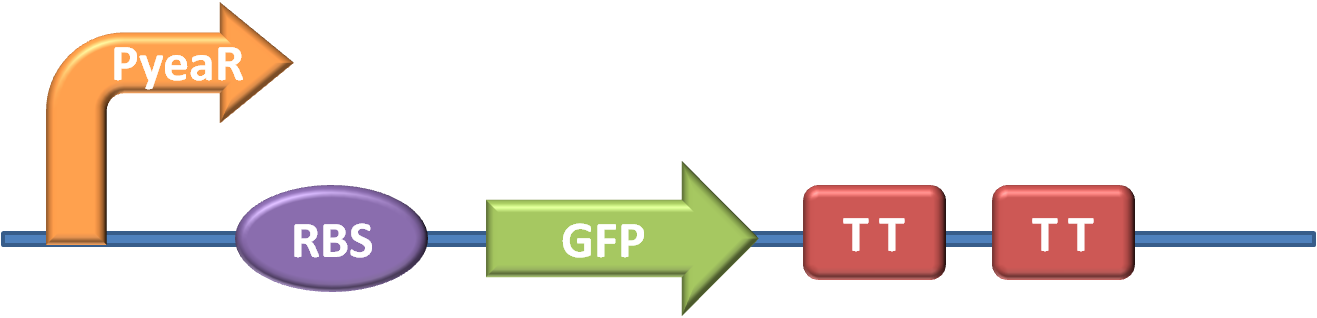
| + | The first stage in the project was to acquire the necessary parts and construct our key BioBrick - one that would trigger GFP production in response to Nitrates or Nitrites. The design of the part can be seen in fig. 1 (for more detailed information about our new brick BBa_K381001 look on our part design page) | ||
| + | |||
| + | The promoter is BBa_K216005, and was requested from the parts registry | ||
| + | The GFP generator along with strong RBS and transcription terminators is BBa_E0840, and was taken from well 12O kitplate 1 of the 2010 distribution. | ||
| + | |||
| + | [[Image:Part.png|centre|500px]] | ||
| + | |||
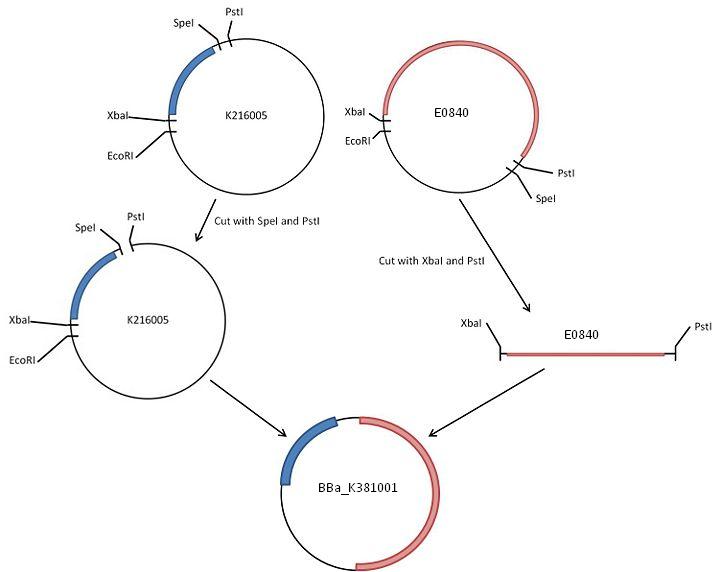
| + | These two parts were cut from their respective plasmids and ligated together. The resulting part was then transformed into commercial NovaBlue cells, then miniprepped to generate large amounts. | ||
| + | |||
| + | Construction of the BioBrick can be broadly broken down into | ||
| + | * Generating larger quantities of both our component BioBricks | ||
| + | * Digesting each of these BioBricks | ||
* Phosphatase treatment of the vector and gel extraction of the digests | * Phosphatase treatment of the vector and gel extraction of the digests | ||
| - | * Ligating together the desired parts to create a new | + | * Ligating together the desired parts to create a new BioBrick |
| - | * Generating larger quantities of this | + | * Generating larger quantities of this BioBrick, checking and sequencing |
| + | ==Generating large quantities of DNA== | ||
| - | + | Before using the BioBricks from the parts registry, we first had to obtain large amounts of these plasmids as the amount provided by the registry was is not sufficient. This is done as follows: | |
| - | * | + | |
| + | * Commercial NovaBlue competent cells were transformed with the DNA from the parts registry, typically transforming 50μL of cells with 2μL of DNA | ||
* Colonies from these transformations were then selected and used to grow overnights - using a wire loop to inoculate 5mL of LB medium (5μL of Ampicillin also added) | * Colonies from these transformations were then selected and used to grow overnights - using a wire loop to inoculate 5mL of LB medium (5μL of Ampicillin also added) | ||
* Combined 3mL of BBa_K216005, BBa_E0840 and BBa_E0240 cell cultures in 1.5mL eppendorf tubes twice - total of 6mL combined cell culture for each biobrick. Performed a miniprep on the combined cultures using a Qiagen "QIAprep Spin Miniprep Kit". Eluted 100μL of solution containing BBa_K216005, BBa_E0840 and BBa_E0240 biobricks in high concentration. | * Combined 3mL of BBa_K216005, BBa_E0840 and BBa_E0240 cell cultures in 1.5mL eppendorf tubes twice - total of 6mL combined cell culture for each biobrick. Performed a miniprep on the combined cultures using a Qiagen "QIAprep Spin Miniprep Kit". Eluted 100μL of solution containing BBa_K216005, BBa_E0840 and BBa_E0240 biobricks in high concentration. | ||
| - | Digesting Component | + | ==Digesting Component BioBricks== |
| + | |||
| + | [[Image:New bb.JPG|thumbnail|right|500px]] | ||
| + | |||
| + | Once we possessed large quantities of each of the BioBricks we wanted to use, we then needed to digest them with appropriate enzymes to leave sticky ends that would match correctly, this was done as follows: | ||
* BBa_E0840 and BBa_E0240 were digested with Xba1 and Pst1 using the following mix: | * BBa_E0840 and BBa_E0240 were digested with Xba1 and Pst1 using the following mix: | ||
| Line 26: | Line 43: | ||
** 1.3μL Xba1 | ** 1.3μL Xba1 | ||
** 12.2μL ddH2O | ** 12.2μL ddH2O | ||
| + | |||
*BBa_K216005 was digested with Spe1 and Pst1 using the following mix: | *BBa_K216005 was digested with Spe1 and Pst1 using the following mix: | ||
| Line 34: | Line 52: | ||
** 1μL Spe1 | ** 1μL Spe1 | ||
** 12.2μL ddH2O | ** 12.2μL ddH2O | ||
| + | |||
*Each digest was left for 2 hours at 37°C | *Each digest was left for 2 hours at 37°C | ||
| + | |||
| + | ==Phosphatase treatment== | ||
| + | |||
| + | To prevent self-ligation it was necessary to dephosphorylate the vector (the cut BBa_K216005), to do this: | ||
| + | |||
| + | Mix: | ||
| + | * 50μL BBa_K216005 digestion mix | ||
| + | * 7μL Alkaline Phosphatase buffer 10x | ||
| + | * 7μL Alkaline Phosphatase | ||
| + | * 6μL ddH2O | ||
| + | |||
| + | Incubate at 37°C for 1 hour | ||
| + | |||
| + | ==Running Gel and Extraction== | ||
| + | |||
| + | To acquire the fragments of DNA that we wanted both the BBa_E0840 digestion mix and BBa_K216005 phosphorylated mix were mixed with 6x loading buffer and run on an agarose gel for 2 hours at 100V. | ||
| + | |||
| + | A Quiagen gel extraction kit was then used to separate our desired fragments, following the procedure supplied with the kit. | ||
| + | |||
| + | ==Ligation== | ||
| + | |||
| + | With our fragments extracted, we then ligated the pieces together to create our new BioBrick, using the following mix: | ||
| + | |||
| + | * 2μL vector (BBa_K216005) | ||
| + | * 6μL insert (BBa_E0840) | ||
| + | * 2μL 10x T4 ligase buffer | ||
| + | * 1μL T4 ligase | ||
| + | * 9μL ddH20 | ||
| + | |||
| + | A control was also made up without any insert and the ligation mix and control were then placed in an eppendorf holder in an ice bucket. This bucket was then left on our desk overnight, allowing the ice to melt and the temperature the ligation mix was at to gradually rice. | ||
| + | |||
| + | ==Transformation== | ||
| + | |||
| + | Our ligation and control mixes were then used to transform commercial NovaBlue cells. 5μL of ligation mix/ control was mixed with 30μL of competent cells and NovaBlue transformation protocol followed. These cells were then spread on plates with the appropriate antibiotic and left overnight at 37°C. | ||
| + | |||
| + | The result was 306 colonies on the plate with the ligation mix and 1 colony on the control plate, indicating a probable success. | ||
| + | |||
| + | ==Acquiring Large Amounts of DNA== | ||
| + | |||
| + | To generate large amounts of our DNA, a colony from the successful plate was used to grow an overnight of cells hopefully transformed with BBa_K381001. This overnight was then used to perform a miniprep, obtaining 50μL of the new biobrick. | ||
| + | |||
| + | To check the construction was successful, a sample from the miniprep was then cut with Xba1 and Spe1 and the result run on a gel. This generated fragments of the size we had expected, which combined with a positive GFP assay indicated our construction had indeed worked. | ||
Latest revision as of 19:06, 27 October 2010
iGEM 2010
Constructing Our Biobrick - K381001
Overview
Contents |
The promoter is BBa_K216005, and was requested from the parts registry The GFP generator along with strong RBS and transcription terminators is BBa_E0840, and was taken from well 12O kitplate 1 of the 2010 distribution.
These two parts were cut from their respective plasmids and ligated together. The resulting part was then transformed into commercial NovaBlue cells, then miniprepped to generate large amounts.
Construction of the BioBrick can be broadly broken down into
- Generating larger quantities of both our component BioBricks
- Digesting each of these BioBricks
- Phosphatase treatment of the vector and gel extraction of the digests
- Ligating together the desired parts to create a new BioBrick
- Generating larger quantities of this BioBrick, checking and sequencing
Generating large quantities of DNA
Before using the BioBricks from the parts registry, we first had to obtain large amounts of these plasmids as the amount provided by the registry was is not sufficient. This is done as follows:
- Commercial NovaBlue competent cells were transformed with the DNA from the parts registry, typically transforming 50μL of cells with 2μL of DNA
- Colonies from these transformations were then selected and used to grow overnights - using a wire loop to inoculate 5mL of LB medium (5μL of Ampicillin also added)
- Combined 3mL of BBa_K216005, BBa_E0840 and BBa_E0240 cell cultures in 1.5mL eppendorf tubes twice - total of 6mL combined cell culture for each biobrick. Performed a miniprep on the combined cultures using a Qiagen "QIAprep Spin Miniprep Kit". Eluted 100μL of solution containing BBa_K216005, BBa_E0840 and BBa_E0240 biobricks in high concentration.
Digesting Component BioBricks
Once we possessed large quantities of each of the BioBricks we wanted to use, we then needed to digest them with appropriate enzymes to leave sticky ends that would match correctly, this was done as follows:
- BBa_E0840 and BBa_E0240 were digested with Xba1 and Pst1 using the following mix:
- 30μL DNA
- 0.5μL BSA 100x
- 5μL NEB Buffer 3
- 1μL Pst1
- 1.3μL Xba1
- 12.2μL ddH2O
- BBa_K216005 was digested with Spe1 and Pst1 using the following mix:
- 30μL DNA
- 0.5μL BSA 100x
- 5μL NEB Buffer 2
- 1.3μL Pst1
- 1μL Spe1
- 12.2μL ddH2O
- Each digest was left for 2 hours at 37°C
Phosphatase treatment
To prevent self-ligation it was necessary to dephosphorylate the vector (the cut BBa_K216005), to do this:
Mix:
- 50μL BBa_K216005 digestion mix
- 7μL Alkaline Phosphatase buffer 10x
- 7μL Alkaline Phosphatase
- 6μL ddH2O
Incubate at 37°C for 1 hour
Running Gel and Extraction
To acquire the fragments of DNA that we wanted both the BBa_E0840 digestion mix and BBa_K216005 phosphorylated mix were mixed with 6x loading buffer and run on an agarose gel for 2 hours at 100V.
A Quiagen gel extraction kit was then used to separate our desired fragments, following the procedure supplied with the kit.
Ligation
With our fragments extracted, we then ligated the pieces together to create our new BioBrick, using the following mix:
- 2μL vector (BBa_K216005)
- 6μL insert (BBa_E0840)
- 2μL 10x T4 ligase buffer
- 1μL T4 ligase
- 9μL ddH20
A control was also made up without any insert and the ligation mix and control were then placed in an eppendorf holder in an ice bucket. This bucket was then left on our desk overnight, allowing the ice to melt and the temperature the ligation mix was at to gradually rice.
Transformation
Our ligation and control mixes were then used to transform commercial NovaBlue cells. 5μL of ligation mix/ control was mixed with 30μL of competent cells and NovaBlue transformation protocol followed. These cells were then spread on plates with the appropriate antibiotic and left overnight at 37°C.
The result was 306 colonies on the plate with the ligation mix and 1 colony on the control plate, indicating a probable success.
Acquiring Large Amounts of DNA
To generate large amounts of our DNA, a colony from the successful plate was used to grow an overnight of cells hopefully transformed with BBa_K381001. This overnight was then used to perform a miniprep, obtaining 50μL of the new biobrick.
To check the construction was successful, a sample from the miniprep was then cut with Xba1 and Spe1 and the result run on a gel. This generated fragments of the size we had expected, which combined with a positive GFP assay indicated our construction had indeed worked.
 "
"