Team:Newcastle/7 July 2010
From 2010.igem.org
Swoodhouse (Talk | contribs) (→Materials and Procedure) |
Swoodhouse (Talk | contribs) (→Chromosomal prep) |
||
| (2 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Newcastle/mainbanner}} | {{Team:Newcastle/mainbanner}} | ||
| - | = | + | =PCR to test chromosomal prep= |
==Aim== | ==Aim== | ||
| Line 11: | Line 11: | ||
==Results== | ==Results== | ||
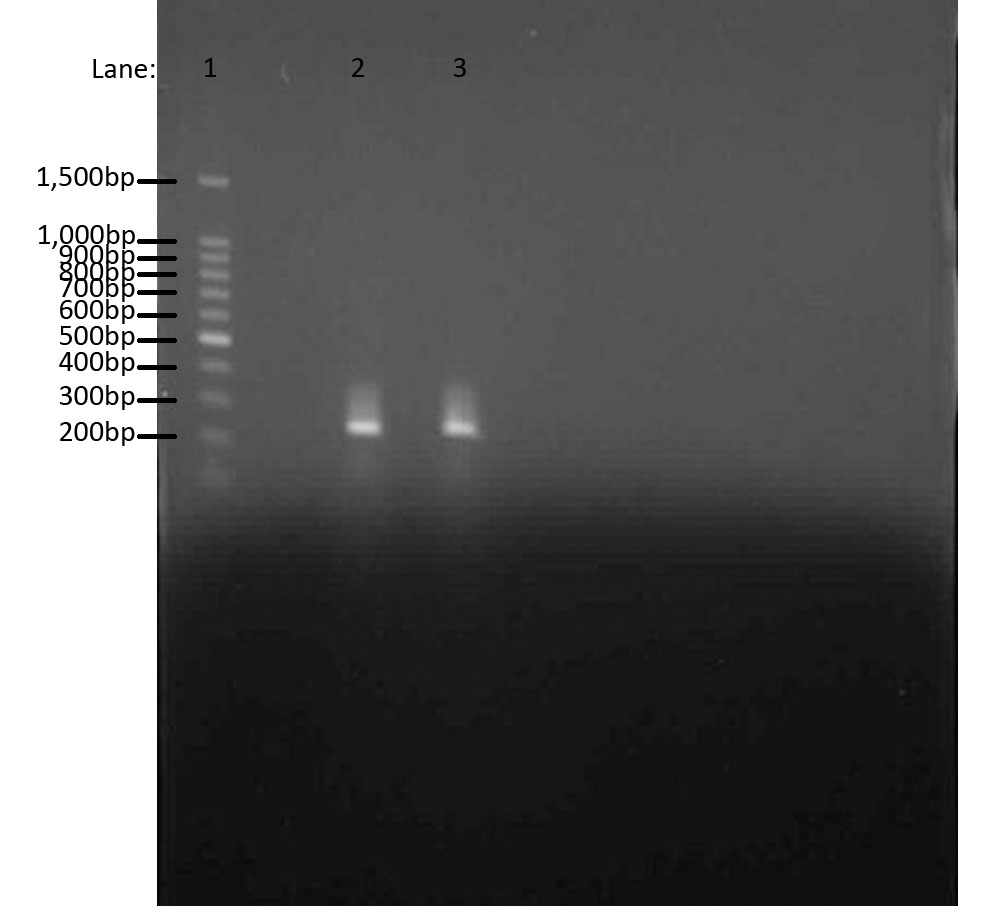
| - | We used | + | We used gel electrophoresis to confirm the quality of our DNA. As we already know, the region our primers amplify is about 300bps. We used 100 bp DNA ladder as a guidance for our bands. The two bands produced were from two separate chromosomal preps. |
[[Image:Newcastle_Genomic_extraction.jpg|400px]] | [[Image:Newcastle_Genomic_extraction.jpg|400px]] | ||
Latest revision as of 15:09, 27 October 2010

| |||||||||||||
| |||||||||||||
Contents |
PCR to test chromosomal prep
Aim
To test whether the chromosome extraction from Bacillus subtilis ATCC6633 was successful.
Materials and Procedure
Please refer to: PCR and Gel electrophoresis for materials and protocol. We used ara forward and reverse primers for amplification.
Results
We used gel electrophoresis to confirm the quality of our DNA. As we already know, the region our primers amplify is about 300bps. We used 100 bp DNA ladder as a guidance for our bands. The two bands produced were from two separate chromosomal preps.
- Lane 1: 100 bp DNA ladder
- Lane 2: Chromosomal DNA with ara gene 1
- Lane 3: Chromosomal DNA with ara gene 2
Conclusion
The aim of this whole experiment was to extract genomic DNA from Bacillus subtilis ATCC 6633. In order to test whether we had extracted the DNA, we first used PCR to amplify and then used Gel Electrophoresis to compare the bands of ara genes to the 100 bp ladder. It worked!
 
|
 "
"