Team:RMIT Australia/Modeling
From 2010.igem.org
(→Taq DNA Polymerase) |
(→Differences between Wildtype and Redesigned Taq) |
||
| (29 intermediate revisions not shown) | |||
| Line 408: | Line 408: | ||
= Modelling = | = Modelling = | ||
| - | == | + | == Parameters used to compare the Models == |
| - | + | [[Image:Table.jpg]] | |
| - | + | == Explanation of Parameters == | |
| + | Using a proteomics server, in our case ExPASy we determined the aliphatic and instability index, extinction coefficient and theoretical PI which will help us distinguish which of the mutations would be best to increase the thermo stability of the TAQ. The isoelectric point (PI) is the pH at which the protein carries no net charge. When the theoretical PI is reached the protein has no net charge hence being immobile in an electric field. From table 1 it is evident that there is no large difference between the theoretical isoelectric points of the original Taq to the Taq with the mutations. The estimated stability of the Taq in the test tube is provided by the instability index. From table 1 we see that the instability index is stable as the predicted numbers are below 40, as anything above this will show an unstable protein. The relative volume occupied by aliphatic side chains of the following amino acids; alanine, valine, isolecine and leucine. The aliphatic index of the protein is regarded as a positive factor for the increase of the thermostability, From Table 1 we see that in both the wild type and the redesigned Taq’s aliphatic indexes were increased as the mutations were made thus indicating that these mutations increased the thermostability of this protein. Using the proteomics server the extinction coefficient were measured as it indicates how much light a protein absorbs at a certain wavelength and is useful later when we purify the protein. | ||
| - | + | ==Taq Polymerase== | |
| - | + | === Taq Polymerase with no mutations === | |
| - | |||
| - | + | <br> | |
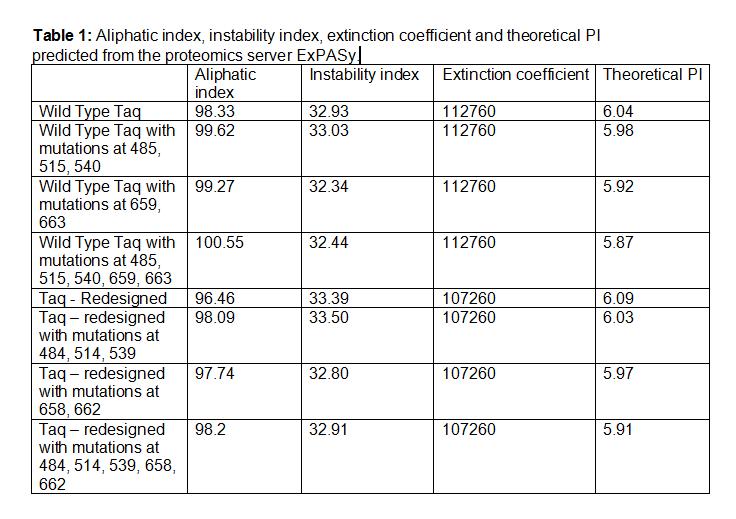
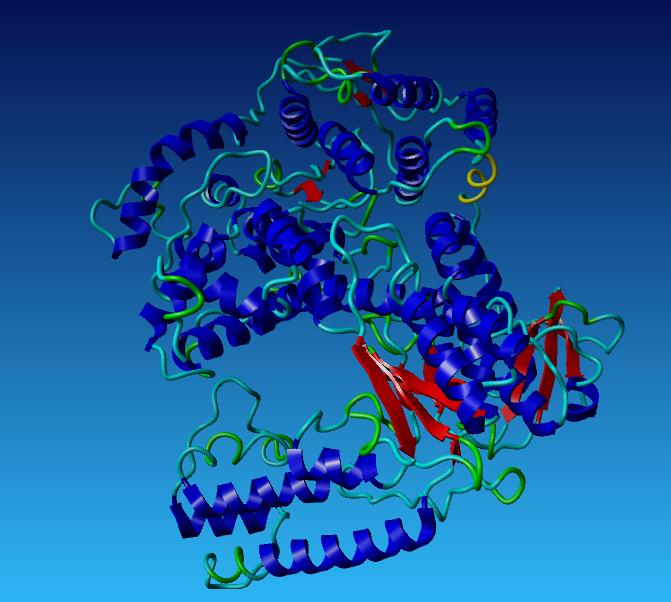
| + | [[Image:WildTypetaq.jpg|400px]] [[Image:wildtype2.jpg|393px]] | ||
| + | <br> | ||
| + | *'''Aliphatic Index:''' 98.33 | ||
| + | |||
| + | *'''Ext Coefficient:''' 112760 | ||
| + | |||
| + | *'''Theoretical PI:''' 6.04 | ||
| + | |||
| + | *'''Instability Index:''' 32.93 | ||
---- | ---- | ||
| + | |||
| + | <br> | ||
| + | |||
| + | === Redesigned Taq (MutTaq)=== | ||
| + | <br> | ||
| + | We redesigned TAQ so it is suitable to undergo ligation independent cloning, to do this we changed the N and C terminal. | ||
| + | <br> | ||
| + | This is the redesigned Taq Polymerase | ||
| + | |||
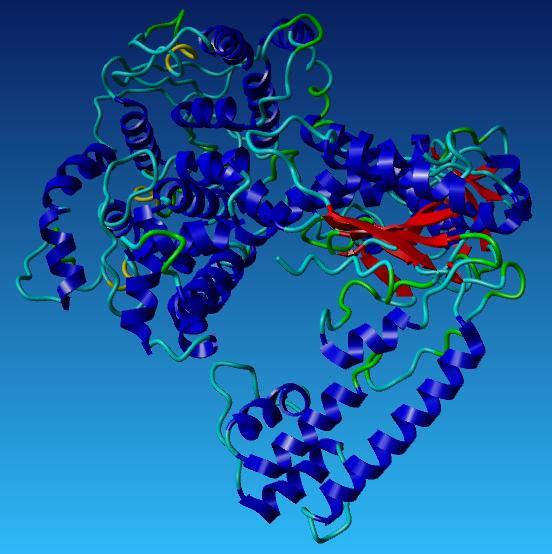
| + | [[Image:Taq part 1.jpg|400px]] [[Image:Taq part 2.jpg|400px]] | ||
| + | |||
| + | |||
| + | *'''Aliphatic Index:''' 96.46 | ||
| + | |||
| + | *'''Ext Coefficient:''' 107260 | ||
| + | |||
| + | *'''Theoretical PI:''' 6.09 | ||
| + | |||
| + | *'''Instability Index:''' 33.39 | ||
| + | |||
| + | |||
| + | The wild type Taq Polymerase was changed at the N and C terminal, with the Root Mean Square Deviation of 6.8834. This number indicates the measure of the average distance between the backbones of the two superimposed proteins. | ||
| + | |||
| + | ----- | ||
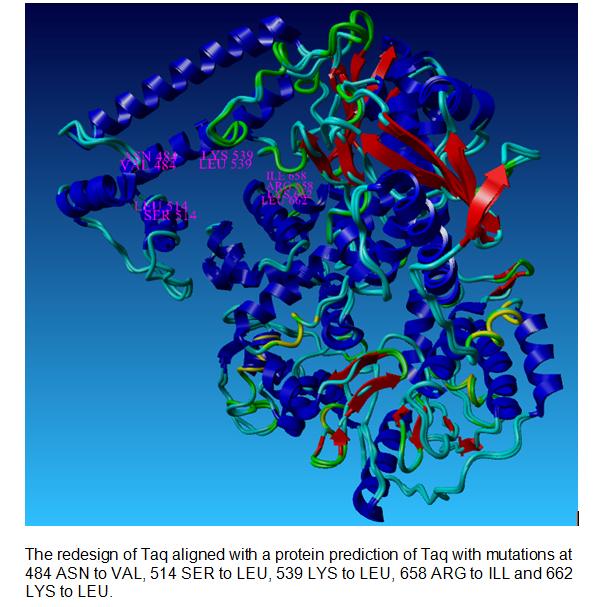
| + | ===Our Taq Polymerase with mutations=== | ||
| + | |||
| + | [[Image:Taq with labels.jpg|centre]] | ||
| + | |||
| + | |||
| + | ===Differences between Wildtype and Redesigned Taq=== | ||
| + | |||
| + | '''The picture below shows the differences between the wild type Taq Polymerase and the redesigned Taq at the N and C terminus.''' | ||
| + | |||
| + | |||
| + | [[Image:Taq N.jpg|530px|centre]] | ||
| + | |||
| + | [[Image:Taq C.jpg|500px|centre]] | ||
== Fold-it== | == Fold-it== | ||
Latest revision as of 09:53, 27 October 2010


Modelling
Parameters used to compare the Models
Explanation of Parameters
Using a proteomics server, in our case ExPASy we determined the aliphatic and instability index, extinction coefficient and theoretical PI which will help us distinguish which of the mutations would be best to increase the thermo stability of the TAQ. The isoelectric point (PI) is the pH at which the protein carries no net charge. When the theoretical PI is reached the protein has no net charge hence being immobile in an electric field. From table 1 it is evident that there is no large difference between the theoretical isoelectric points of the original Taq to the Taq with the mutations. The estimated stability of the Taq in the test tube is provided by the instability index. From table 1 we see that the instability index is stable as the predicted numbers are below 40, as anything above this will show an unstable protein. The relative volume occupied by aliphatic side chains of the following amino acids; alanine, valine, isolecine and leucine. The aliphatic index of the protein is regarded as a positive factor for the increase of the thermostability, From Table 1 we see that in both the wild type and the redesigned Taq’s aliphatic indexes were increased as the mutations were made thus indicating that these mutations increased the thermostability of this protein. Using the proteomics server the extinction coefficient were measured as it indicates how much light a protein absorbs at a certain wavelength and is useful later when we purify the protein.
Taq Polymerase
Taq Polymerase with no mutations
- Aliphatic Index: 98.33
- Ext Coefficient: 112760
- Theoretical PI: 6.04
- Instability Index: 32.93
Redesigned Taq (MutTaq)
We redesigned TAQ so it is suitable to undergo ligation independent cloning, to do this we changed the N and C terminal.
This is the redesigned Taq Polymerase
- Aliphatic Index: 96.46
- Ext Coefficient: 107260
- Theoretical PI: 6.09
- Instability Index: 33.39
The wild type Taq Polymerase was changed at the N and C terminal, with the Root Mean Square Deviation of 6.8834. This number indicates the measure of the average distance between the backbones of the two superimposed proteins.
Our Taq Polymerase with mutations
Differences between Wildtype and Redesigned Taq
The picture below shows the differences between the wild type Taq Polymerase and the redesigned Taq at the N and C terminus.
Fold-it

Foldit is a revolutionary new computer game enabling you to contribute to important scientific research. This page describes the science behind Foldit and how your playing can help.
[http://fold.it/portal/ Click Here]
 "
"