Team:HokkaidoU Japan/Notebook/August13
From 2010.igem.org
(Difference between revisions)
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{Template:HokkaidoU_Japan}} | + | {{Template:HokkaidoU_Japan}}<div class="linkbar"><div class="prev">[[Team:HokkaidoU_Japan/Notebook/August12|August 12]]</div>[[Team:HokkaidoU_Japan/Notebook|Notebook]]<div class="next">[[Team:HokkaidoU_Japan/Notebook/August16|August 16]]</div></div> |
| - | = | + | |
| - | = | + | =PCR of parts which didn't amplify well via mini prep= |
| - | == | + | ==Composition of Reaction Solution== |
| - | {| | + | {|class="protocol" |
| + | |- | ||
| + | !Reagent | ||
| + | !Amount | ||
|- | |- | ||
|style="text-align:right;"| Autoclaved DW : | |style="text-align:right;"| Autoclaved DW : | ||
| Line 26: | Line 29: | ||
|style="text-align:left;"| 1 uL | |style="text-align:left;"| 1 uL | ||
|- | |- | ||
| - | |style="text-align:right;"| Template (1-18F) : | + | |style="text-align:right;"| Template ([[Team:HokkaidoU_Japan/Parts#BioBricks|1-18F]]) : |
|style="text-align:left;"| 1 uL | |style="text-align:left;"| 1 uL | ||
|- | |- | ||
| - | |style="text-align:right; border-top:1px solid # | + | |style="text-align:right; border-top:1px solid #996;"|'''Total''' : |
| - | |style="text-align:left; border-top:1px solid # | + | |style="text-align:left; border-top:1px solid #996;"| 50 uL |
|} | |} | ||
| - | == | + | ==Removal of Primers== |
| - | # | + | # Added 150 uL of TE to 50 uL of PCR product, each |
| - | # | + | # Transfered into Microcon YM-10 filter and cetrifuged for 20 min at 14,000 G |
| - | # | + | # Much of the solution remained so centrifuged for aditional 10 min |
| - | # | + | # Measured the amount left |
| - | # | + | ## It was 140 uL so centrifuged again for 10min |
| + | # And again | ||
| + | # Finally DNA solution was reduced to 19 uL so we added 31 uL to make final volume of 50 uL | ||
| + | |||
| + | ==Electrophoresis== | ||
| + | |||
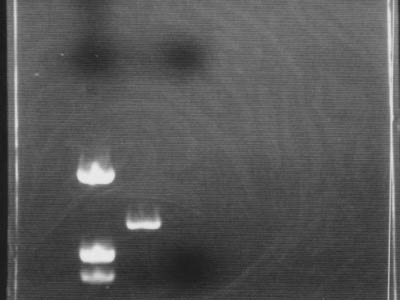
| + | [[Image:HokkaidoU Japan 20100813a.jpg|200px|right|thumb|PCR for DNA confirmation]] | ||
| - | + | # Added 0.4 uL of 6x Sample Buffer to 1 uL of DNA solution and electrophoresed it. | |
| - | + | # At the same time add added 2 uL of 6x Sample Buffer to 10 uL of flow trough and centrifuged | |
| - | # | + | * Used marker [https://2010.igem.org/Image:HokkaidoU_Pictures_DNA_Marker.png pUC119/''Hin''f] |
| - | # | + | * It was confirmed that DNA was amplified by PCR |
| - | * | + | ** Between marker bands of 543 bp and 1330 bp PCR product band of 700 bp was visible |
| - | * | + | * Due to negligence (electrophoresis for 45 min), flow through with primers flowed through and exited gel |
| - | * | + | |
<div style="clear:both"> | <div style="clear:both"> | ||
Latest revision as of 07:26, 27 October 2010
PCR of parts which didn't amplify well via mini prep
Composition of Reaction Solution
| Reagent | Amount |
|---|---|
| Autoclaved DW : | 33 uL |
| 10x PCR buffer : | 5 uL |
| 2 mM dNTPs : | 5 uL |
| 25 mM MgSO4 : | 3 uL |
| EX-F primer : | 1 uL |
| PS-R primer : | 1 uL |
| KOD plus Neo : | 1 uL |
| Template (1-18F) : | 1 uL |
| Total : | 50 uL |
Removal of Primers
- Added 150 uL of TE to 50 uL of PCR product, each
- Transfered into Microcon YM-10 filter and cetrifuged for 20 min at 14,000 G
- Much of the solution remained so centrifuged for aditional 10 min
- Measured the amount left
- It was 140 uL so centrifuged again for 10min
- And again
- Finally DNA solution was reduced to 19 uL so we added 31 uL to make final volume of 50 uL
Electrophoresis
- Added 0.4 uL of 6x Sample Buffer to 1 uL of DNA solution and electrophoresed it.
- At the same time add added 2 uL of 6x Sample Buffer to 10 uL of flow trough and centrifuged
- Used marker pUC119/Hinf
- It was confirmed that DNA was amplified by PCR
- Between marker bands of 543 bp and 1330 bp PCR product band of 700 bp was visible
- Due to negligence (electrophoresis for 45 min), flow through with primers flowed through and exited gel
 "
"