Team:HokkaidoU Japan/Notebook/August10
From 2010.igem.org
(Difference between revisions)
| (26 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:HokkaidoU_Japan}} | {{Template:HokkaidoU_Japan}} | ||
| - | <div class="linkbar"><div class="prev">[[Team:HokkaidoU_Japan/Notebook/ | + | <div class="linkbar"><div class="prev">[[Team:HokkaidoU_Japan/Notebook/August2|August 2]]</div>[[Team:HokkaidoU_Japan/Notebook|Notebook]]<div class="next">[[Team:HokkaidoU_Japan/Notebook/August11|August 11]]</div></div> |
| - | == | + | ==Single Colony Isolation== |
| - | + | * Observed if any colonies were made | |
| - | + | ** Could not see [[Team:HokkaidoU_Japan/Parts#BioBricks|3-1E]] | |
| - | * | + | * Number of other colonies on other plates was also very small |
| - | ** | + | ** Mistake in protocol is inferred |
| - | ** | + | ** Precipitation of cells maybe at fault |
| - | * | + | * All but [[Team:HokkaidoU_Japan/Parts#BioBricks|3-1E]] was moved to new plates |
| - | 3- | + | |
| - | == | + | ==[[Team:HokkaidoU_Japan/Protocols|PCR]] Reaction System== |
| - | + | ||
| - | {|style="text-align: center; margin-left:20px" | + | Reaction system was prepared for 3 parts, 245 uL in total |
| + | |||
| + | {|style="text-align: center; margin-left:20px" class="protocol" | ||
| + | |- | ||
| + | !Reagent | ||
| + | !Amount | ||
|- | |- | ||
| Autoclaved DW | | Autoclaved DW | ||
| Line 36: | Line 40: | ||
| 5 uL | | 5 uL | ||
|- | |- | ||
| - | |style="border-top:1px solid # | + | |style="border-top:1px solid #996;"|'''Total''' |
| - | |style="border-top:1px solid # | + | |style="border-top:1px solid #996;"|'''245 uL''' |
|} | |} | ||
| - | * | + | * 49 uL of reaction solution was added to each of 4 tubes and after DNA template was added |
| - | * | + | * Length of each template is listed int the table below |
| - | {|border="1" style="margin-left: 20px;" | + | |
| + | {|border="1" style="margin-left: 20px;" class="protocol" | ||
| + | |- | ||
| + | !Tube | ||
| + | !Biobrick | ||
| + | !Length | ||
|- | |- | ||
| 1 | | 1 | ||
| - | | 2- | + | | [[Team:HokkaidoU_Japan/Parts#BioBricks|2-24G]](sender) |
| 847 bp | | 847 bp | ||
|- | |- | ||
| 2 | | 2 | ||
| - | | 1- | + | | [[Team:HokkaidoU_Japan/Parts#BioBricks|1-2M]](RBS) |
| 61 bp | | 61 bp | ||
|- | |- | ||
| 3 | | 3 | ||
| - | | 1- | + | | [[Team:HokkaidoU_Japan/Parts#BioBricks|1-23L]](terminator) |
| 178 bp | | 178 bp | ||
|- | |- | ||
| 4 | | 4 | ||
| - | | 3- | + | | [[Team:HokkaidoU_Japan/Parts#BioBricks|3-1E]](heat sensor) |
| 984 bp | | 984 bp | ||
|} | |} | ||
| - | * DNA | + | * Template DNA length is calculated by adding prefix and suffix length |
| - | * 3- | + | ** Biobrick Length + Prefix length + Suffix Length |
| + | * Because [[Team:HokkaidoU_Japan/Protocols|Mini prep]] of 3-1E failed, it amplified by PCR | ||
==PCR program== | ==PCR program== | ||
| - | {| | + | {|style="margin-left: 20px;" class="protocol" |
|- | |- | ||
| Predenature | | Predenature | ||
| 94℃ 2 min | | 94℃ 2 min | ||
|- | |- | ||
| - | | Denature | + | |style="border-top:1px solid #996;"| Denature |
| - | | 98℃ 10 sec | + | |style="border-top:1px solid #996;"| 98℃ 10 sec |
|- | |- | ||
| Extension | | Extension | ||
| 68℃ 30 sec | | 68℃ 30 sec | ||
| + | |- | ||
| + | |style="border-top:1px solid #996;"| Hold | ||
| + | |style="border-top:1px solid #996;"| 4℃ | ||
|} | |} | ||
| - | * 30 | + | * 30 cycles |
| - | * 30 sec/ | + | * KOD potency is 30 sec/kbp, so 30 sec for 1 cycle |
| - | == | + | ==Electrophoresis== |
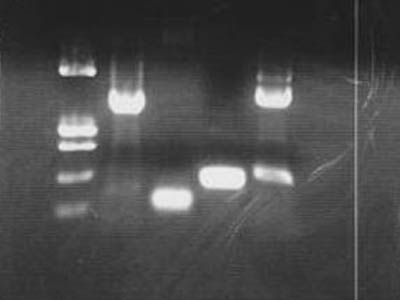
| - | * | + | [[Image:HokkaidoU Japan 20100810a.jpg|200px|right|thumb|Electrophoresis of amplified BioBricks]] |
| - | * | + | |
| - | * | + | * Added 1 uL of 6xSample Buffer to 5 uL of amplified DNA |
| - | [[ | + | * Used marker [https://2010.igem.org/Image:HokkaidoU_Pictures_DNA_Marker.png pUC119/''Hin''f1] |
| + | * Added 20 uL of EtBr to 1/2 TBE | ||
| + | |||
| + | {|border="1" style="margin-left: 20px;" class="protocol" | ||
| + | !Lane | ||
| + | !DNA | ||
| + | !Length of DNA | ||
| + | |- | ||
| + | | 1 | ||
| + | | pUC119/''Hin''f1 | ||
| + | | | ||
| + | |- | ||
| + | | 2 | ||
| + | | [[Team:HokkaidoU_Japan/Parts#BioBricks|2-24G]](sender) | ||
| + | | 847 bp | ||
| + | |- | ||
| + | | 3 | ||
| + | | [[Team:HokkaidoU_Japan/Parts#BioBricks|1-2M]](RBS) | ||
| + | | 61 bp | ||
| + | |- | ||
| + | | 4 | ||
| + | | [[Team:HokkaidoU_Japan/Parts#BioBricks|1-23L]](terminator) | ||
| + | | 178 bp | ||
| + | |- | ||
| + | | 5 | ||
| + | | [[Team:HokkaidoU_Japan/Parts#BioBricks|3-1E]](heat sensor) | ||
| + | | 984 bp | ||
| + | |} | ||
Latest revision as of 07:14, 27 October 2010
Single Colony Isolation
- Observed if any colonies were made
- Could not see 3-1E
- Number of other colonies on other plates was also very small
- Mistake in protocol is inferred
- Precipitation of cells maybe at fault
- All but 3-1E was moved to new plates
PCR Reaction System
Reaction system was prepared for 3 parts, 245 uL in total
| Reagent | Amount |
|---|---|
| Autoclaved DW | 165 uL |
| 10x PCR buffer | 25 uL |
| 2 mM dNTPs | 25 uL |
| 25 mM MgSO4 | 15 uL |
| EX-F primer | 5 uL |
| PS-R primer | 5 uL |
| KOD plus Neo | 5 uL |
| Total | 245 uL |
- 49 uL of reaction solution was added to each of 4 tubes and after DNA template was added
- Length of each template is listed int the table below
| Tube | Biobrick | Length |
|---|---|---|
| 1 | 2-24G(sender) | 847 bp |
| 2 | 1-2M(RBS) | 61 bp |
| 3 | 1-23L(terminator) | 178 bp |
| 4 | 3-1E(heat sensor) | 984 bp |
- Template DNA length is calculated by adding prefix and suffix length
- Biobrick Length + Prefix length + Suffix Length
- Because Mini prep of 3-1E failed, it amplified by PCR
PCR program
| Predenature | 94℃ 2 min |
| Denature | 98℃ 10 sec |
| Extension | 68℃ 30 sec |
| Hold | 4℃ |
- 30 cycles
- KOD potency is 30 sec/kbp, so 30 sec for 1 cycle
Electrophoresis
- Added 1 uL of 6xSample Buffer to 5 uL of amplified DNA
- Used marker pUC119/Hinf1
- Added 20 uL of EtBr to 1/2 TBE
| Lane | DNA | Length of DNA |
|---|---|---|
| 1 | pUC119/Hinf1 | |
| 2 | 2-24G(sender) | 847 bp |
| 3 | 1-2M(RBS) | 61 bp |
| 4 | 1-23L(terminator) | 178 bp |
| 5 | 3-1E(heat sensor) | 984 bp |
 "
"