Team:Heidelberg/Notebook/miRNA Kit/August
From 2010.igem.org
Laura Nadine (Talk | contribs) |
(→24/08/2010) |
||
| (17 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single}} |
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single_Pagetop|note_mirna_kit}} |
| - | = 21/08/2010 = | + | {{:Team:Heidelberg/Side_Top}} |
| - | [[ | + | |
| - | [[Image: | + | {| cellpadding="5" cellspacing="0" align="center" style="text-align: center; color:#009be1; border: 1.5px solid #000000;" |
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#f09600;" | [https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August<font color="white">August</font>] | ||
| + | |- style="background:#f09600; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |colspan="6"| ||'''1''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''2'''||'''3'''||'''4'''||'''5'''||'''6'''||'''7'''||'''8''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''9'''||'''10'''||'''11'''||'''12'''||'''13'''||'''14'''||'''15''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''16'''||'''17'''||'''18'''||'''19'''||'''20'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#21/08/2010 21]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#22.2F08.2F2010 22]''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#23.2F08.2F2010 23]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#24.2F08.2F2010 24]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#25.2F08.2F2010 25]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#26.2F08.2F2010 26]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#27.2F08.2F2010 27]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#28.2F08.2F2010 28]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#29.2F08.2F2010 29]''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''[[Igem2010/Main/synthetic_miR_Kit/August#30/08/2010|30]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/August#31/08/2010|31]]'''||colspan="5"| | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| cellpadding="5" cellspacing="0" align="center" style="text-align: center; color:#78b41e; border: 1.5px solid #333333;" | ||
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#009be1;" | [https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September<font color="#ffecba">September</font>] | ||
| + | |- style="background:#009be1; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |colspan="2"| ||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#01.2F09.2F2010 1]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#02.2F09.2F2010 2]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#03.2F09.2F2010 3]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#04.2F09.2F2010 4]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#05.2F09.2F2010 5]''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#06.2F09.2F2010 6]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#07.2F09.2F2010 7]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#08.2F09.2F2010 8]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#09.2F09.2F2010 9]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#10.2F09.2F2010 10]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#11.2F09.2F2010 11]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#12.2F09.2F2010 12]''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#13.2F09.2F2010 13]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#14.2F09.2F2010 14]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#15.2F09.2F2010 15]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#16.2F09.2F2010 16]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#17.2F09.2F2010 17]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#18.2F09.2F2010 18]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#19.2F09.2F2010 19]''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#20.2F09.2F2010 20]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#21.2F09.2F2010 21]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#22.2F09.2F2010 22]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#23.2F09.2F2010 23]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#24.2F09.2F2010 24]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#25.2F09.2F2010 25]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#26.2F09.2F2010 26]''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#27.2F09.2F2010 27]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#28.2F09.2F2010 28]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#29.2F09.2F2010 29]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#22.2F30.2F2010 30]'''||colspan="3"| | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | | colspan="7"| <span style="color:#ffffff">-</span> | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| cellpadding="5" cellspacing="0" align="center" style="text-align: center; color:#f09600; border: 1.5px solid #333333;" | ||
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#78b41e;" | [https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October<font color="white">October</font>] | ||
| + | |- style="background:#78b41e; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |colspan="4"| ||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#01.2F10.2F2010 1]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#02.2F10.2F2010 2]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#03.2F10.2F2010 3]''' | ||
| + | |- style="background:#f2f2f2; color:#f09600" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#04.2F10.2F2010 4]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#05.2F10.2F2010 5]''' | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#06.2F10.2F2010 6]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#07.2F10.2F2010 7]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#08.2F10.2F2010 8]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#09.2F10.2F2010 9]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#10.2F10.2F2010 10]''' | ||
| + | |- style="background:#f2f2f2; color:#f09600" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#11.2F10.2F2010 11]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#12.2F10.2F2010 12]'''|'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#13.2F10.2F2010 13]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#14.2F10.2F2010 14]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#15.2F09.2F2010 15]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#16.2F10.2F2010 16]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#17.2F10.2F2010 17]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#18.2F10.2F2010 18]''' | ||
| + | |- style="background:#f2f2f2; color:#f09600" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#19/10/2010 19]'''||'''20'''||'''21'''||'''22'''||'''23'''||'''24'''||'''25''' | ||
| + | |- style="background:#f2f2f2; color:#f09600" | ||
| + | |'''26'''||'''27'''||'''28'''||'''29'''||'''30'''||colspan="3"| | ||
| + | |- style="background:#f2f2f2; color:#f09600" | ||
| + | | colspan="7"| <span style="color:#ffffff">-</span> | ||
| + | |} | ||
| + | |||
| + | |||
| + | {{:Team:Heidelberg/Side_Bottom}} | ||
| + | |||
| + | __NOTOC__ | ||
| + | |||
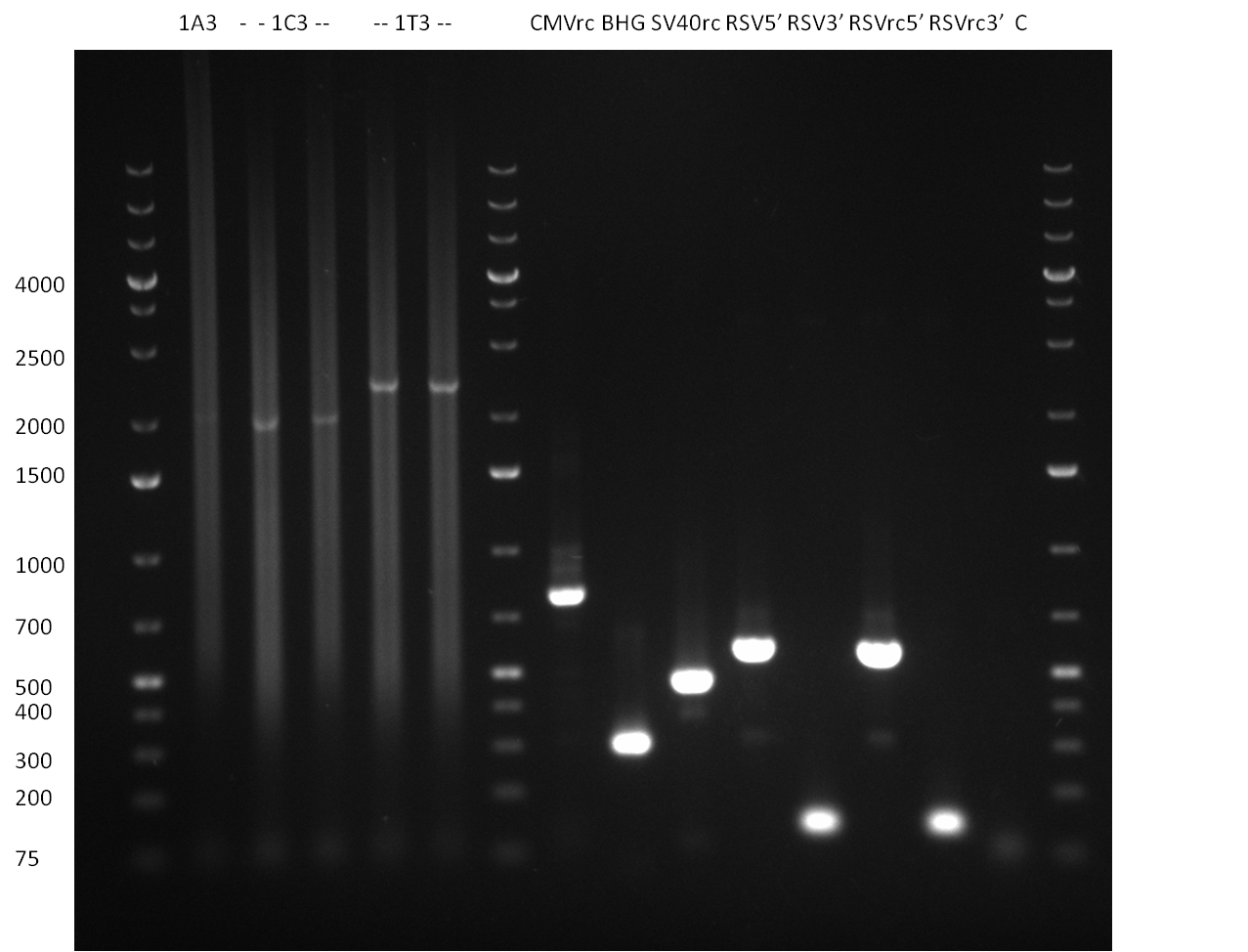
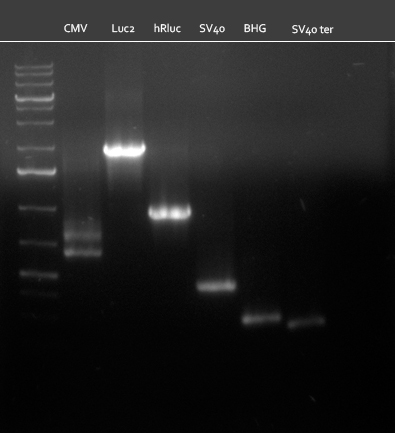
| + | ===21/08/2010=== | ||
| + | [[Image:20100820_parts.png|thumb|350 px|left|Gel1]] | ||
| + | |||
<br /> | <br /> | ||
* all primers for the synthetic miR construction Kit were diluted to a 100 uM concentration | * all primers for the synthetic miR construction Kit were diluted to a 100 uM concentration | ||
* PCR reaction for construction of the following parts were set up: | * PCR reaction for construction of the following parts were set up: | ||
| + | |||
<br /> | <br /> | ||
| + | [[Image:20100822_gel1.png|thumb|350 px|right|Gel2]] | ||
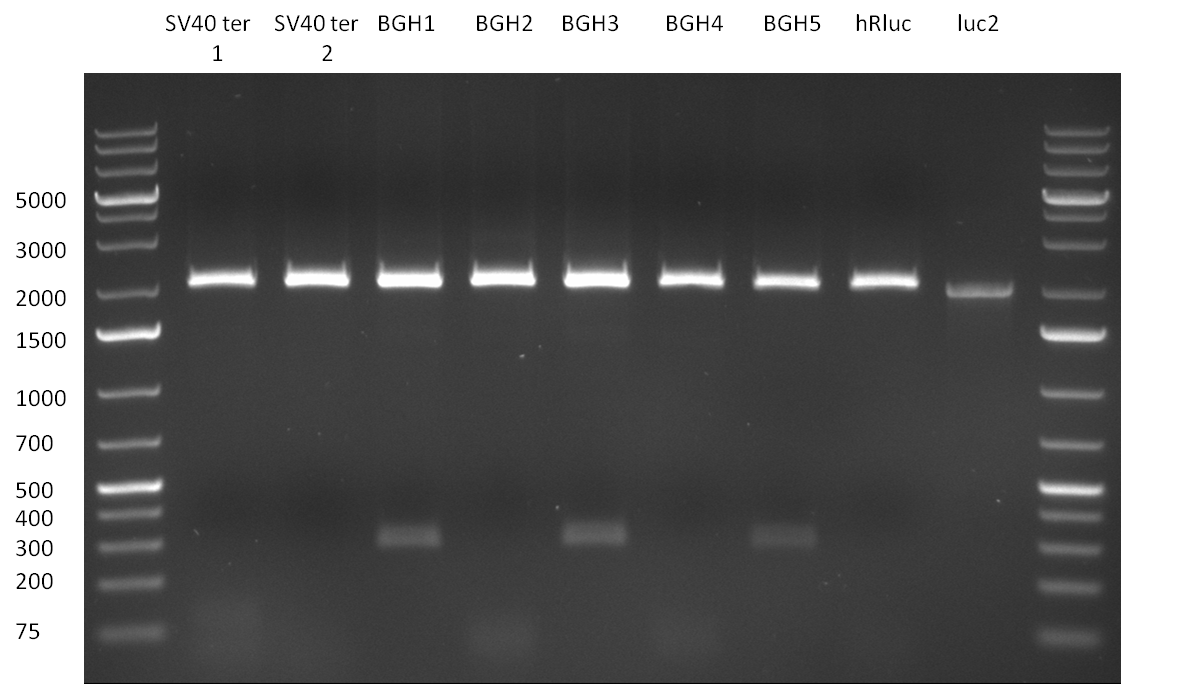
:* CMV (reverse complementary); template: pCMV_BBB, primer: CMV_rc_BBB_fw/rev | :* CMV (reverse complementary); template: pCMV_BBB, primer: CMV_rc_BBB_fw/rev | ||
:* BGH terminator; template: pcDNA5/TO/FRT/miRsAg, primer: BGH_pA_fw/rev | :* BGH terminator; template: pcDNA5/TO/FRT/miRsAg, primer: BGH_pA_fw/rev | ||
| Line 73: | Line 142: | ||
<br /> | <br /> | ||
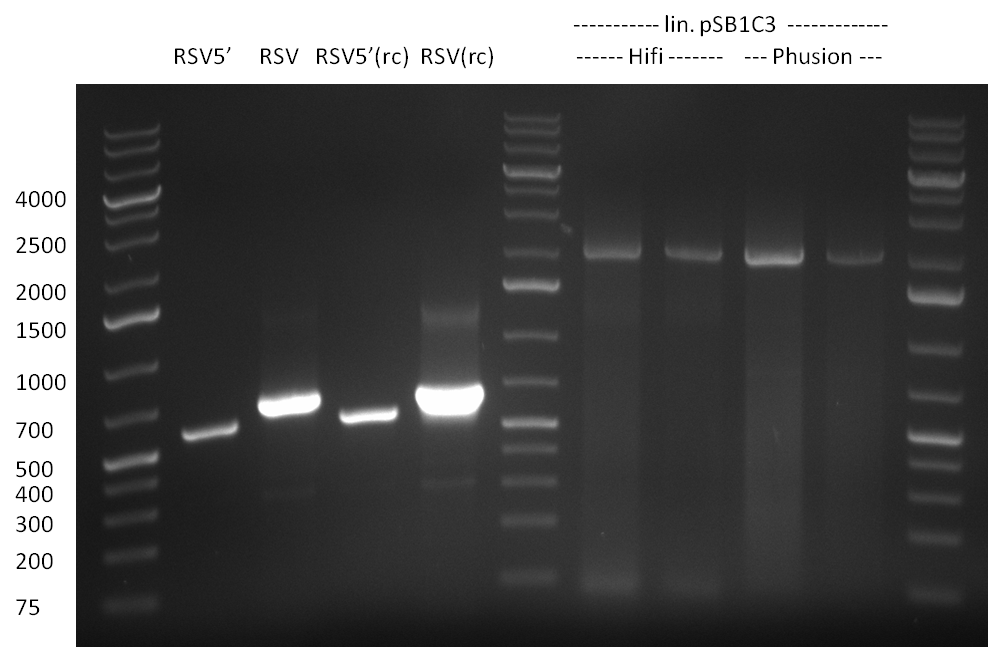
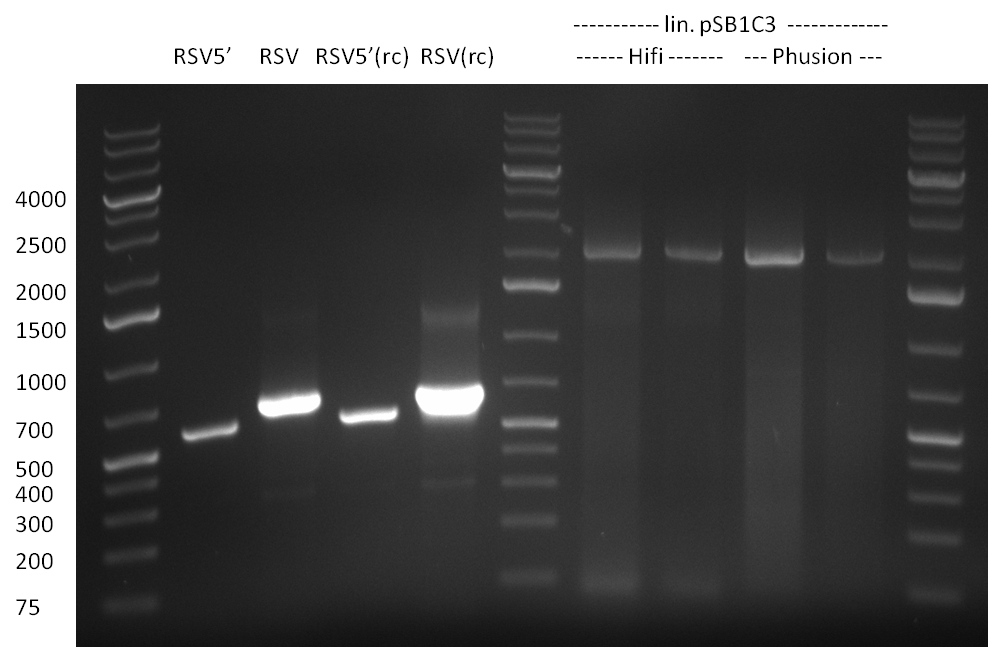
| - | + | ===22/08/2010=== | |
| - | + | [[Image:20100822_gel2.png|thumb|350px|right]] | |
| - | = 22/08/2010 = | + | |
| - | [[Image:20100822_gel2.png|thumb| | + | |
<br /> | <br /> | ||
* Fusion-PCR for the construction of the whole RSV promoter fragment with BBB prefix and suffix: | * Fusion-PCR for the construction of the whole RSV promoter fragment with BBB prefix and suffix: | ||
| Line 131: | Line 198: | ||
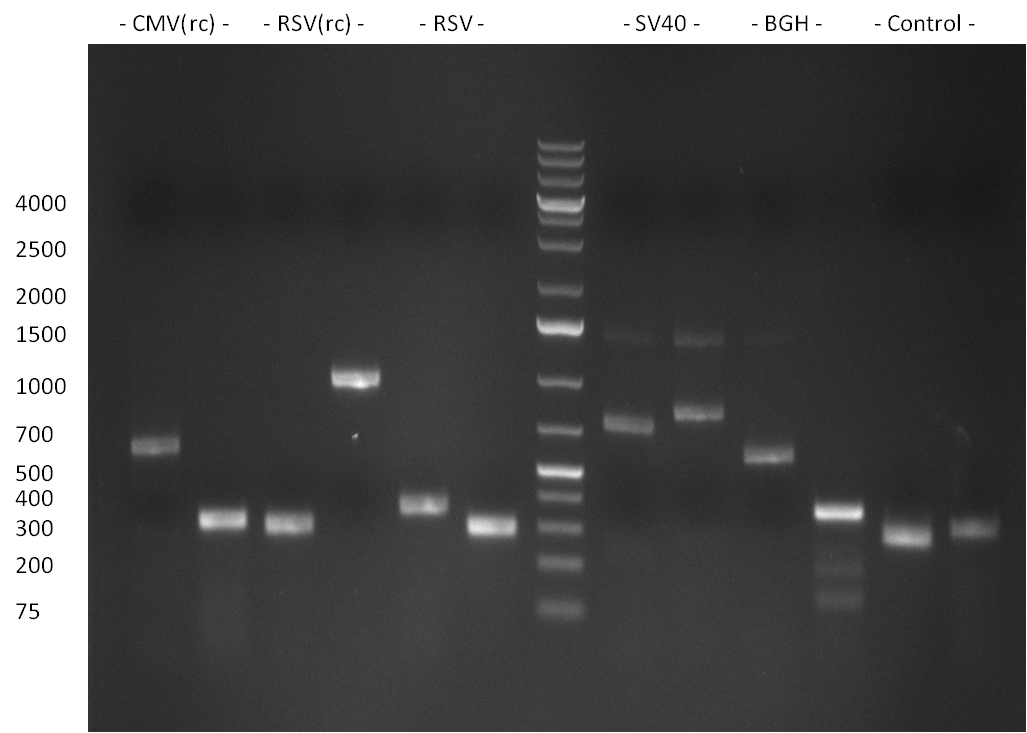
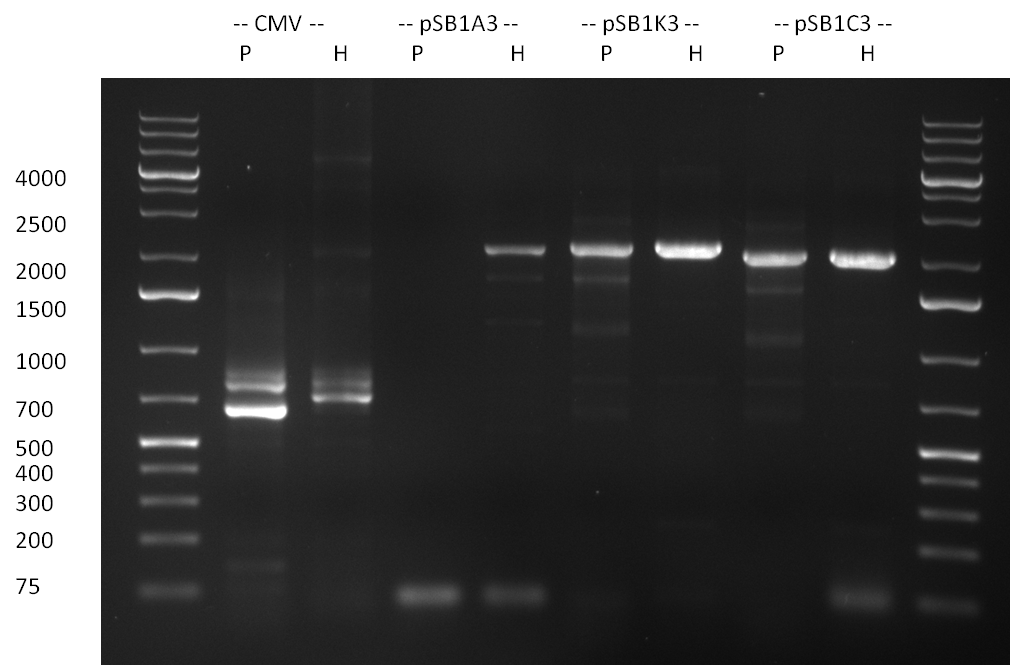
| - | = 23/08/2010 = | + | ===23/08/2010=== |
| - | [[image:20100824_gel1.png|thumb| | + | [[image:20100824_gel1.png|thumb|350px|right|gel1]] |
| - | [[Image:20100823_CMV_Luc2_hRluc_SV40_BGH_SV40ter.jpg|thumb| | + | [[Image:20100823_CMV_Luc2_hRluc_SV40_BGH_SV40ter.jpg|thumb|350px|right|Gel2]] |
<br /> | <br /> | ||
Colony-PCR of the colonies obtained on the agar-plates (cloning previous day) was performed according to the following protocol: | Colony-PCR of the colonies obtained on the agar-plates (cloning previous day) was performed according to the following protocol: | ||
| Line 163: | Line 230: | ||
PCR was performed according to the standard Hifi-Protocol (annealing temperature 57 °C) | PCR was performed according to the standard Hifi-Protocol (annealing temperature 57 °C) | ||
| - | + | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | |
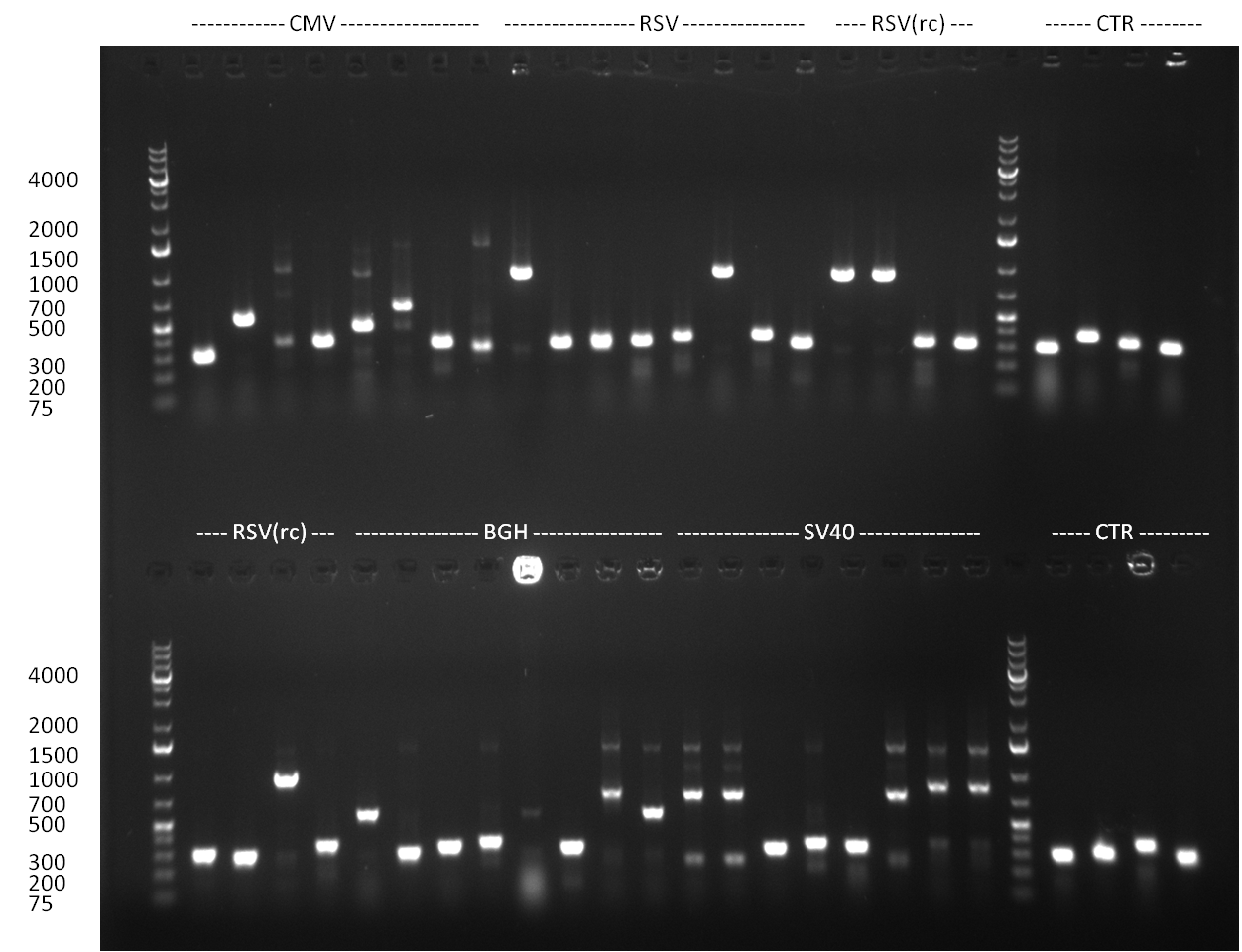
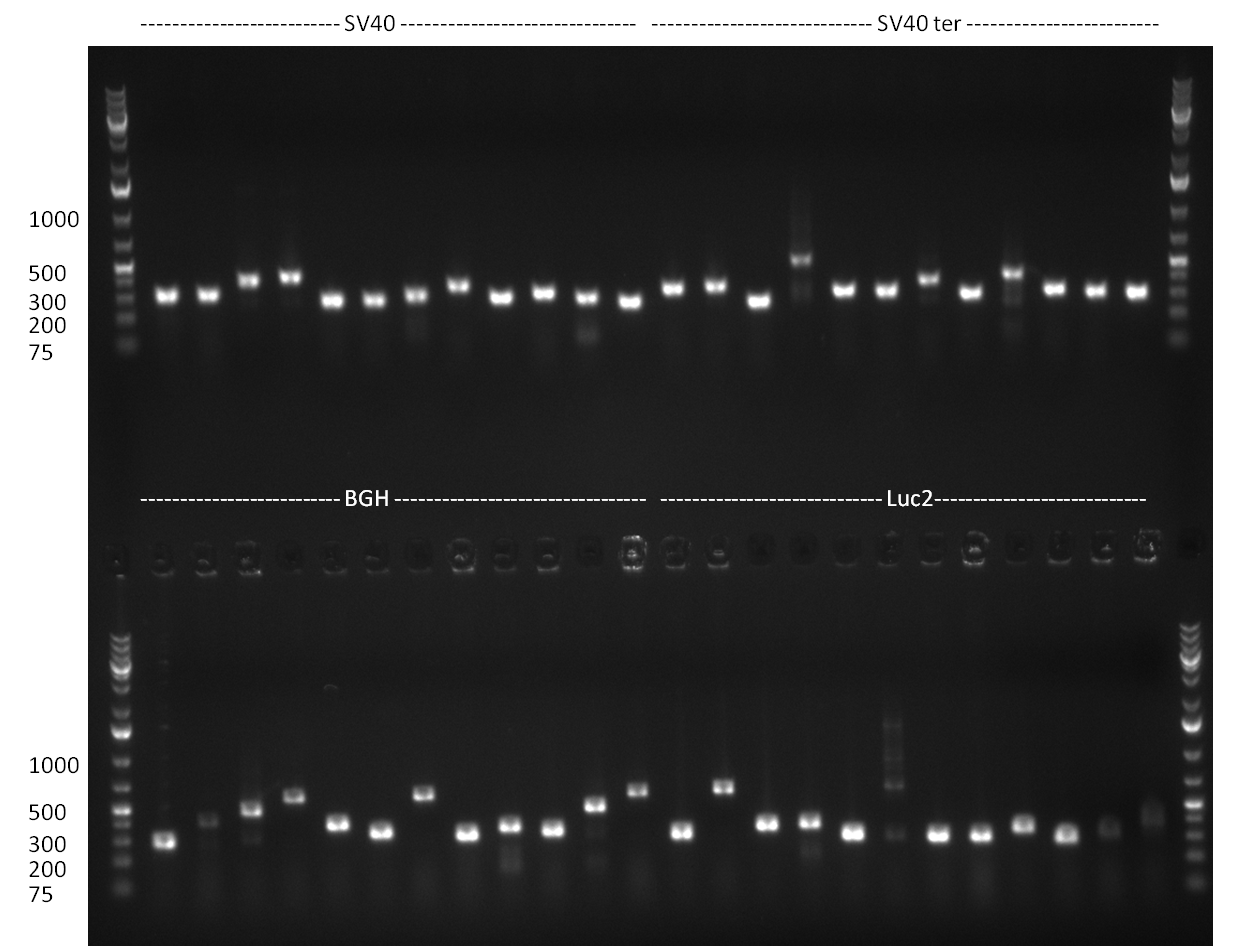
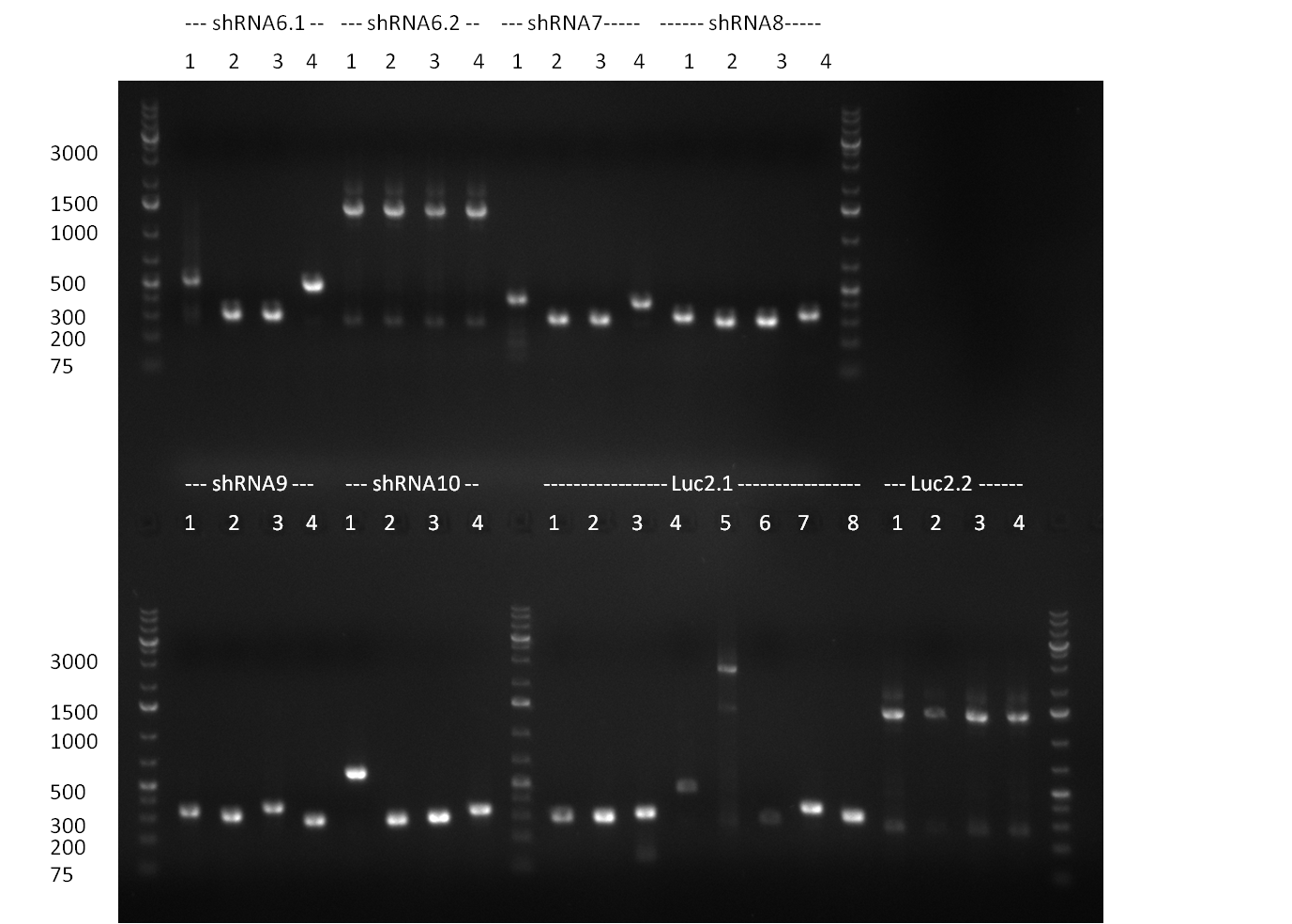
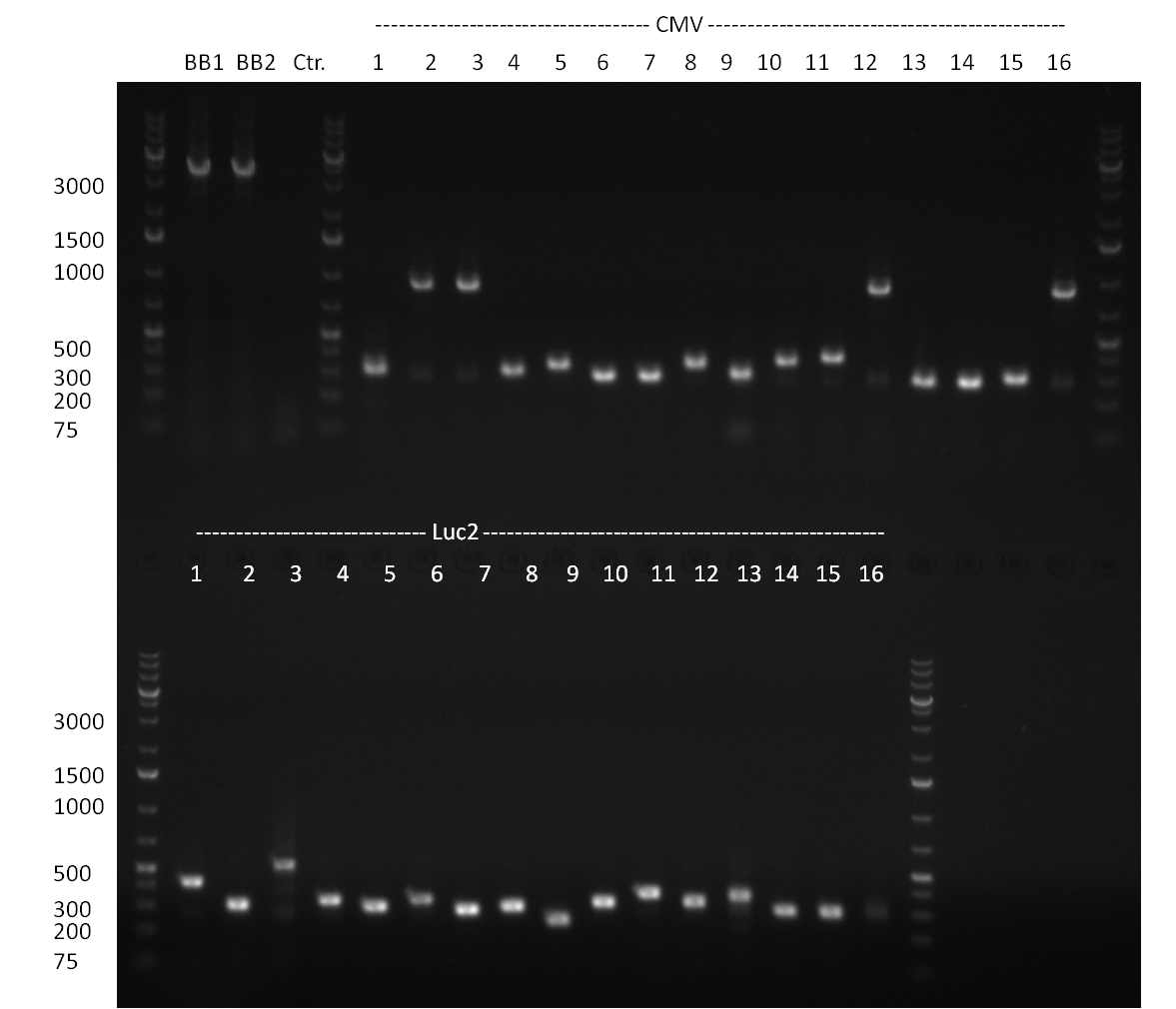
| - | + | ===24/08/2010=== | |
| - | + | [[image: 20100824_gel1_2.png|thumb|350 px|right|Gel1]] | |
| - | + | [[image: 20100824_gel2.png|thumb|350 px|right|Gel2]] | |
| - | + | [[image: 20100824_gel3.png|thumb|350 px|right|Gel3]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | = 24/08/2010 = | + | |
| - | [[image: 20100824_gel1_2.png|thumb| | + | |
| - | [[image: 20100824_gel2.png|thumb| | + | |
| - | [[image: 20100824_gel3.png|thumb| | + | |
<br /> | <br /> | ||
* Amplification of the CMV Promoter as well as the amplification of pSB1A3/C3/K3 standard plasmid backbones was performed via Phusion or Hifi-PCR according to the standard protocol (annealing temperature 61 °C). | * Amplification of the CMV Promoter as well as the amplification of pSB1A3/C3/K3 standard plasmid backbones was performed via Phusion or Hifi-PCR according to the standard protocol (annealing temperature 61 °C). | ||
| Line 221: | Line 271: | ||
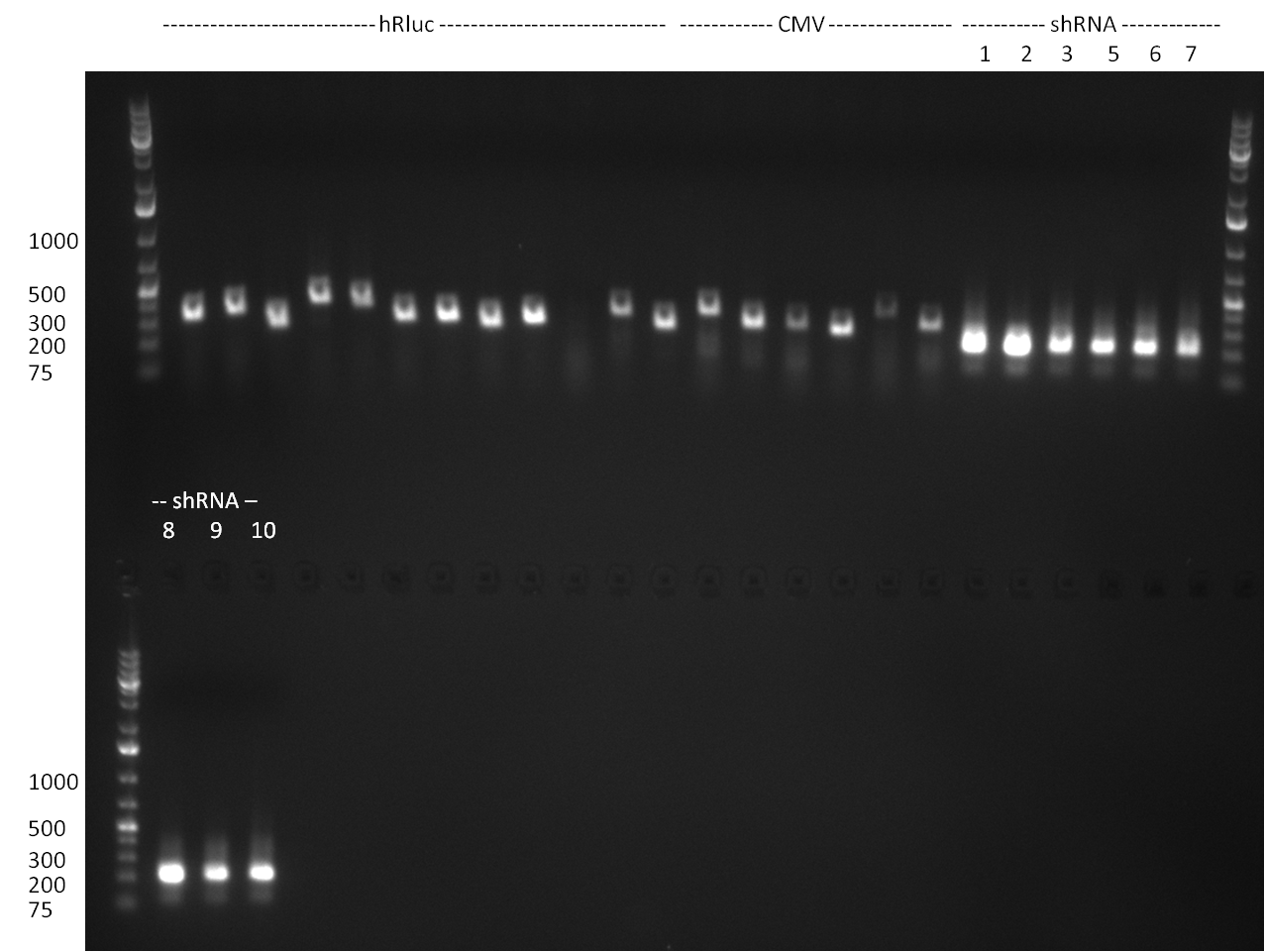
* PCR for the construction of synthetic shRNAs was performed. Therefore, the pcDNA5/TO/FRT containing the shRNA miRsAg as insert was used as template. For the shRNAs 1,2,3,5,6,7,8,9,10 the synthesis of two intermediate fragments were performed. The 5' fragment was amplified by using the primer shRNA_AflI_BBb_fw and the reverse oligos of the certain shRNA gene. The 3' framents were amplified by using the primer shRNA_HindIII_BBb_rev and the forward primer of the certain shRNA gene. The forward and reverse primer of the shRNA gene introduce the guiding and passanger with certain specificities into the shRNA constuct. The miRsAg is an miR-122 like shRNA and uses the backbone of miR-122, but another guiding and passanger strand. The PCR was set up according to the standard protocol (annealing temperature: 58 °C). | * PCR for the construction of synthetic shRNAs was performed. Therefore, the pcDNA5/TO/FRT containing the shRNA miRsAg as insert was used as template. For the shRNAs 1,2,3,5,6,7,8,9,10 the synthesis of two intermediate fragments were performed. The 5' fragment was amplified by using the primer shRNA_AflI_BBb_fw and the reverse oligos of the certain shRNA gene. The 3' framents were amplified by using the primer shRNA_HindIII_BBb_rev and the forward primer of the certain shRNA gene. The forward and reverse primer of the shRNA gene introduce the guiding and passanger with certain specificities into the shRNA constuct. The miRsAg is an miR-122 like shRNA and uses the backbone of miR-122, but another guiding and passanger strand. The PCR was set up according to the standard protocol (annealing temperature: 58 °C). | ||
The results of the PCR were analyzed on a 2 % agarose gel, run for 30 min @ 100 V (gel 3)). | The results of the PCR were analyzed on a 2 % agarose gel, run for 30 min @ 100 V (gel 3)). | ||
| - | + | <br /><br /><br /><br /><br /> | |
| - | = 25/08/2010 = | + | ===25/08/2010=== |
<br /> | <br /> | ||
The constructs were tested in a digestion with NotI using the following protocol: | The constructs were tested in a digestion with NotI using the following protocol: | ||
| Line 243: | Line 293: | ||
| - | = 26/08/2010 = | + | ===26/08/2010=== |
<br /> | <br /> | ||
*The sequencing result for the samples send for sequencing on the privious day was checked. The results were positive for all samples, sequences were accurate. | *The sequencing result for the samples send for sequencing on the privious day was checked. The results were positive for all samples, sequences were accurate. | ||
*Transformation of the ligation done on the previous day according to the standard transformation protocol. | *Transformation of the ligation done on the previous day according to the standard transformation protocol. | ||
<br /> | <br /> | ||
| - | = 27/08/2010 = | + | ===27/08/2010=== |
| - | [[Image:20100827_tet_colonyPCR.png |thumb| | + | [[Image:20100827_tet_colonyPCR.png |thumb|350px|right|gel1]] |
| - | [[Image:20100827_3vectors_digested_EcoPstDpn.png |thumb| | + | [[Image:20100827_3vectors_digested_EcoPstDpn.png |thumb|350px|right|gel2]] |
| Line 309: | Line 359: | ||
<br /> | <br /> | ||
| - | = 28/08/2010 = | + | ===28/08/2010=== |
| - | [[Image:20100828_gel1.png|thumb| | + | [[Image:20100828_gel1.png|thumb|350 px|right]] |
| - | [[Image:20100828_gel2.png|thumb| | + | [[Image:20100828_gel2.png|thumb|350 px|right]] |
<br /> | <br /> | ||
*6 more colonies from the cloning done on the 25th/26th were picked and colony PCR was performed according to the standard colony-PCR protocol. In parallel, Minipreps of each colony were inocculated. The results of the colony-PCR ist shown in the gelpictures. | *6 more colonies from the cloning done on the 25th/26th were picked and colony PCR was performed according to the standard colony-PCR protocol. In parallel, Minipreps of each colony were inocculated. The results of the colony-PCR ist shown in the gelpictures. | ||
| Line 319: | Line 369: | ||
<br /> | <br /> | ||
| - | = 29/08/2010 = | + | ===29/08/2010=== |
| - | [[Image:20100829_gel2.png|thumb| | + | [[Image:20100829_gel2.png|thumb|350px|right|gel1]] |
| - | [[Image:20100829_gel1.png|thumb| | + | [[Image:20100829_gel1.png|thumb|350px|right|gel2]] |
<br /> | <br /> | ||
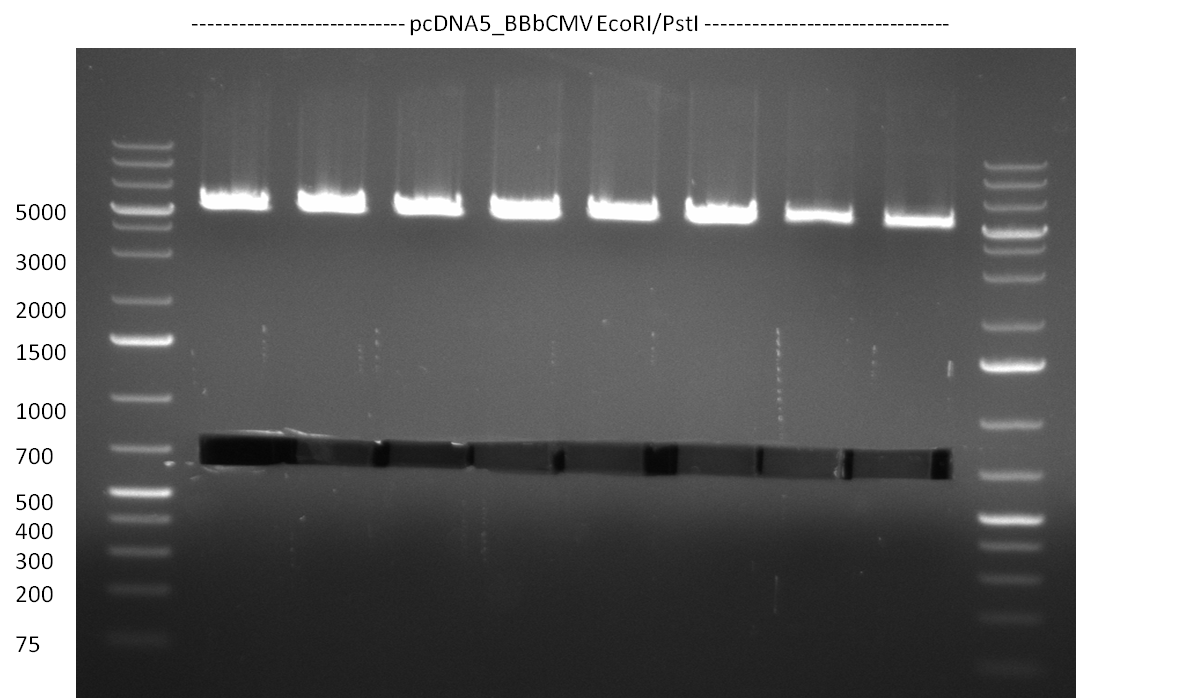
* Vector pcDNA_BBbCMV, PCR product Luc2_ter and shRNA(1-10) with EcoRI/PstI (1 ug each). The BBb_CMV was cut out from the gel (~ 700 bp band) and gel purified (gel1). The PCR product Luc2_ter was purified by applying a nucleotide removal kit. | * Vector pcDNA_BBbCMV, PCR product Luc2_ter and shRNA(1-10) with EcoRI/PstI (1 ug each). The BBb_CMV was cut out from the gel (~ 700 bp band) and gel purified (gel1). The PCR product Luc2_ter was purified by applying a nucleotide removal kit. | ||
| Line 328: | Line 378: | ||
<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
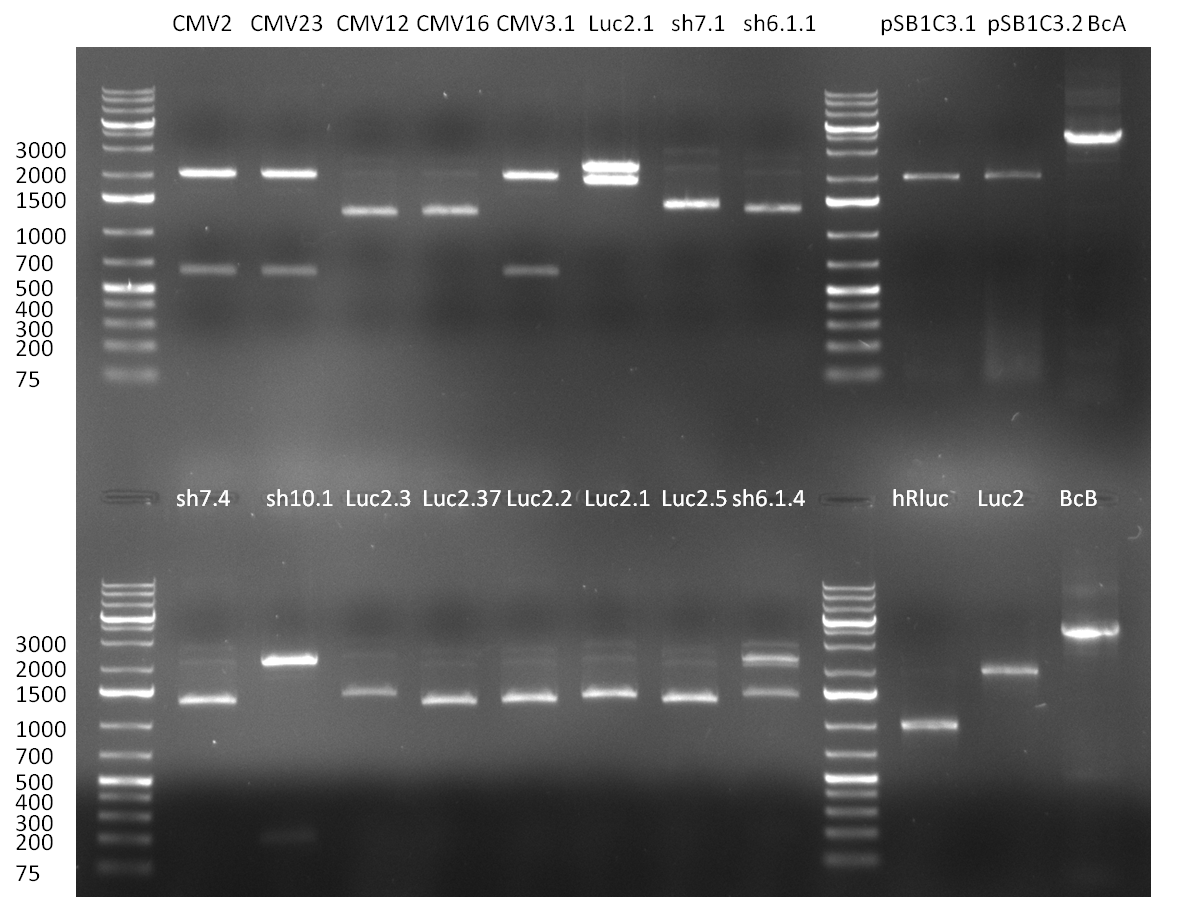
| - | = 30/08/2010 = | + | ===30/08/2010=== |
| - | [[Image:20100830_gel1.png|thumb| | + | [[Image:20100830_gel1.png|thumb|350 px|right|Gel1]] |
| - | [[Image:20100830_gel2.png|thumb| | + | [[Image:20100830_gel2.png|thumb|350 px|right|Gel2]] |
| - | [[Image:20100830_gel3.png|thumb| | + | [[Image:20100830_gel3.png|thumb|350 px|left|Gel3]] |
<br /> | <br /> | ||
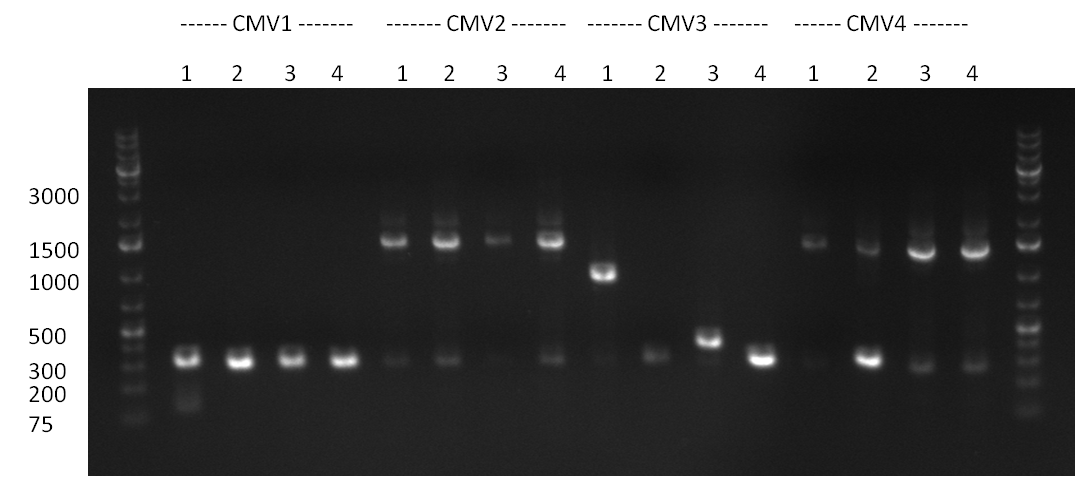
* The ligation result from the previous days' cloning was analyzed via colony PCR (gel 1-3) and all colonies were inocculated (LB cultures) The construct numbers indicate the number of the clone picked from the agar plate. If cloning was performed in Replicates, the first number indicates the plate (i.e. CMV3) and the second number the clone. As seen in gelpicture 1 the clone CMV3.1 and on gelpicture 3 the clones CMV 2,3,12 and 16 all show the right band length of ~1000 bp. On gelpicture 2 the shRNA10.1 clone shows the right band as well as the Luc2.1.5 clone for the Luc2 luciferase. | * The ligation result from the previous days' cloning was analyzed via colony PCR (gel 1-3) and all colonies were inocculated (LB cultures) The construct numbers indicate the number of the clone picked from the agar plate. If cloning was performed in Replicates, the first number indicates the plate (i.e. CMV3) and the second number the clone. As seen in gelpicture 1 the clone CMV3.1 and on gelpicture 3 the clones CMV 2,3,12 and 16 all show the right band length of ~1000 bp. On gelpicture 2 the shRNA10.1 clone shows the right band as well as the Luc2.1.5 clone for the Luc2 luciferase. | ||
| Line 337: | Line 387: | ||
<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
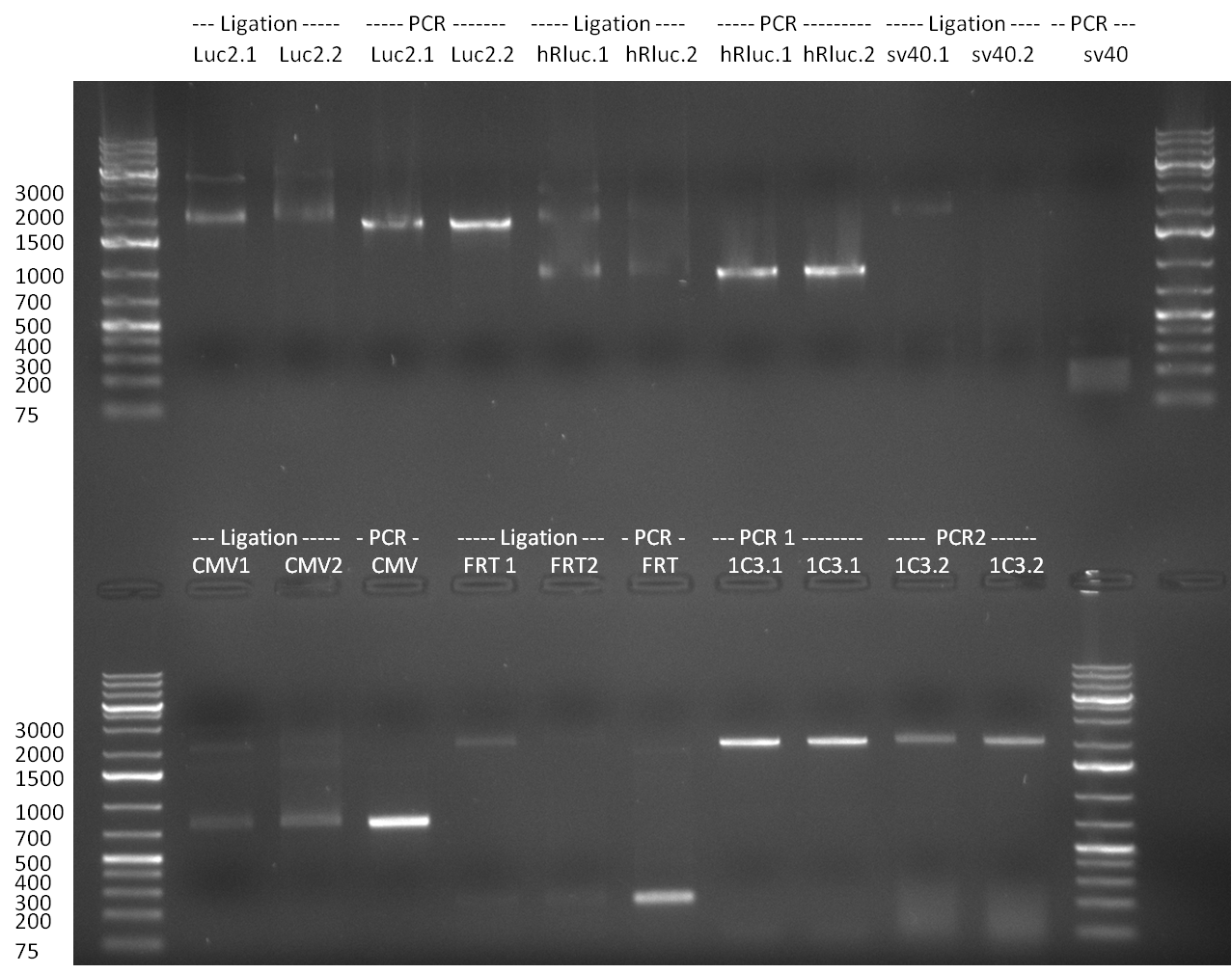
| - | = 31/08/2010 = | + | ===31/08/2010=== |
[[Image:20103108_gel1bb.png|thumb|350 px|right|Gel1]] | [[Image:20103108_gel1bb.png|thumb|350 px|right|Gel1]] | ||
[[Image:20100831_gel2bb.png|thumb|350 px|right|Gel2]] | [[Image:20100831_gel2bb.png|thumb|350 px|right|Gel2]] | ||
| Line 347: | Line 397: | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single_Bottom}} |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 05:32, 27 October 2010

21/08/2010
................................................
................................................ (35x)
................................................
................................................
................................................
................................................ (35x)
................................................
................................................
The 2.1 kb band was afterwards cut out from the gel and a gel extraction was applied for purification of the DNA (Qiagen Gel Extraction Kit).
22/08/2010
................................................
................................................ (35x)
................................................
................................................
The PCR for both fragments was analyzed on a 1 % agarose gel, run for 1 h @100 V (Gel1) and PCR purified applying a Qiagen PCR purification Kit.
23/08/2010
24/08/2010
The PCR product was run on a Gel for 1 h @ 100 V and analyzed an a 1 % agarose gel (gel 1).
The results of the PCR were analyzed on a 2 % agarose gel, run for 30 min @ 100 V (gel 3)).
25/08/2010
Digestion of pSB1C3 and the PCR products of the mentioned constructs with EcoRI/PstI (1 ug each) according to the standard digestion protocol.
Subsequent purification using a nucleaotide removal kit and ligation overnight at 16 °C.
26/08/2010
27/08/2010
1. Backbone amplification using primers backbone_AvrII_fw and backbone_AvrII_rev 2. CMV-TetO2: CMV_TetO2_BBB_fw, CMV_TetO2_BBB_rev 3. Tet-repressor: TetRepressor (from pcDNA6)fw, TetRepressor (from pcDNA6)rew 4. FRT site: FRT_BBB_fw, FRT_BBB_rev
5 ul of each plasmid (conc. = ?) 3 ul EcoRI buffer 3 ul BSA 0.5 ul of each enzyme 18 ul nuclease-free water Incubatation: 37 C, 1hr Heat inactivation: 80 C, 20 min Then DpnI was added (0.5 ul), and the tubes were incubated at 37 C for 1 hr, then heat-inactivation at 80 C for 20 min. 5 ul of the product were loaded on a 1% agarose gel and analyzed (gel1).
28/08/2010
29/08/2010
30/08/2010
31/08/2010
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"