Team:Heidelberg/Project/miRNA Kit
From 2010.igem.org
(→Introduction) |
(→Results) |
||
| Line 34: | Line 34: | ||
==Results== | ==Results== | ||
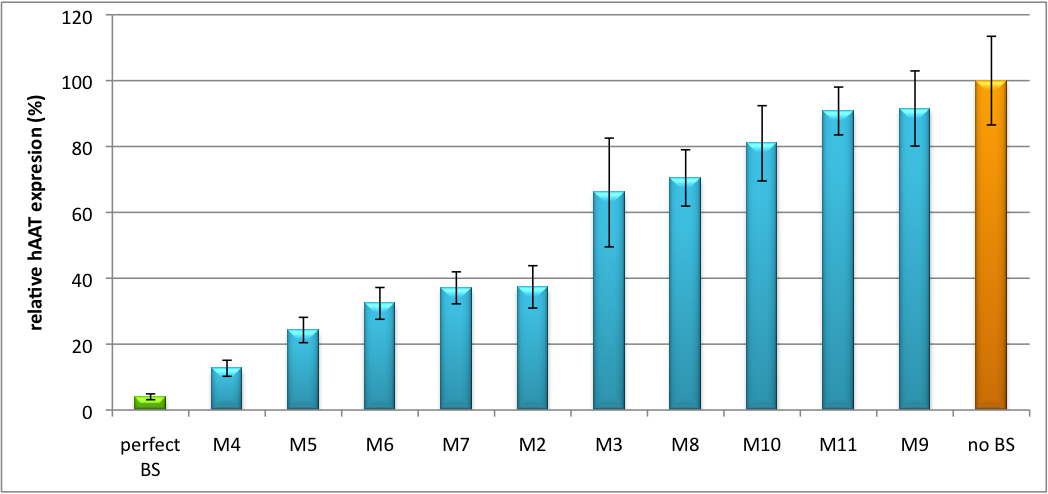
| - | + | Here, we clearly show, that we are able to subsequently tune gene expression over a wide linear range from almost 0% to 100% as compared to perfect and no binding sites. For detailed measurement procedure descriptions, see [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Dual_Luciferase_Assay methods] | |
| + | [[Image:Tuning H1 1.png|thumb|center|600px|'''Figure 1: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_H1''' ...]] | ||
| + | even better: | ||
| + | [[Image:Tuning U6 1.png|thumb|center|600px|'''Figure 1: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_U6''' ...]] | ||
| - | |||
| - | |||
| + | <!--After creating a binding site library and testing the miRNA-binding site interaction <i>in vitro</i>, we were able to compute an [https://2010.igem.org/Team:Heidelberg/Modeling/miGUI <i>in silico</i> model] based on a machine learning approach to predict knockdown efficiencies. A more detailed description of the different binding sites, we characterized can be found in our [https://2010.igem.org/Team:Heidelberg/Project/miMeasure measurements] page. | ||
| - | We further tested our kit using a gene that is an interesting candidate for gene therapy, human alpha-1-antitrypsin (haat) (ref, description). In this approach, we tagged haat, that we used as our GOI, with binding sites that we measured and characterized with our [https://2010.igem.org/Team:Heidelberg/Project/miMeasure miMeasure] construct beforehand. This was a first potential therapeutic approach applying [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#ELISA ELISA] for measurements. | + | Another application of our synthetic miRNA Kit profits of tissue specific endogenous miRNAs expression. These can be exploited for On and Off-Targeting. On targeting in this case would mean that the presence of a certain miRNA in a cell switches on expression of the GOI. This can be accomplished by using a repressor that is targeted by an endogenously expressed miRNA. We exemplified this scenario by using a Tet Repressor fused with a perfect binding site for miRNA 122, a liver-specific miRNA (REF!). At the same time, the promoter expressing the GOI would be under control of a Tet Operator. Upon presence of the miRNA 122, the Tet Repressor would be knocked down, release the promoter and expression of the GOI could be established. |
| + | (YOUC AN CHANGE THIS INTO PAST TENSE IF IT WORKED. AND ADD THE OFF SWITCHE; I AM NOT CERTAIN OF WHAT WE DID THERE, AND FIGURES!) | ||
| + | |||
| + | We further tested our kit using a gene that is an interesting candidate for gene therapy, human alpha-1-antitrypsin (haat) (ref, description). In this approach, we tagged haat, that we used as our GOI, with binding sites that we measured and characterized with our [https://2010.igem.org/Team:Heidelberg/Project/miMeasure miMeasure] construct beforehand. This was a first potential therapeutic approach applying [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#ELISA ELISA] for measurements.--> | ||
<html> | <html> | ||
| Line 48: | Line 53: | ||
</div> | </div> | ||
</html> | </html> | ||
| + | |||
==Discussion== | ==Discussion== | ||
Revision as of 03:17, 27 October 2010

|
|
||
 "
"