Team:Lethbridge/Notebook/Protocols
From 2010.igem.org
Adam.smith4 (Talk | contribs) |
Liszabruder (Talk | contribs) |
||
| (One intermediate revision not shown) | |||
| Line 16: | Line 16: | ||
<th> | <th> | ||
| - | <image src="https://static.igem.org/mediawiki/2010/ | + | <image src="https://static.igem.org/mediawiki/2010/9/91/UofLLabWork.JPG" height="300px"/> |
</th> | </th> | ||
| Line 150: | Line 150: | ||
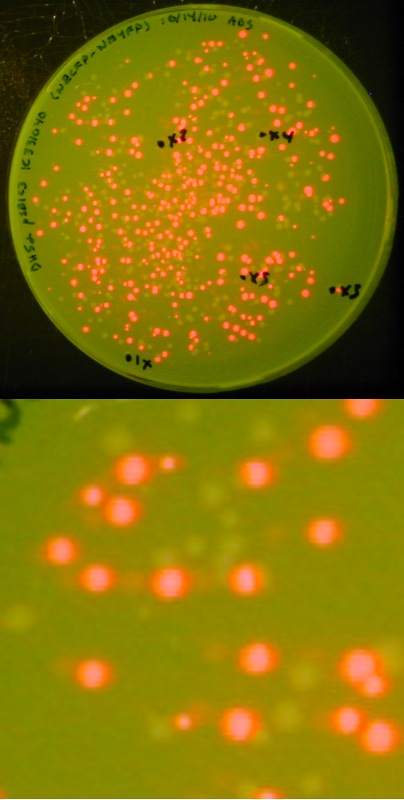
[[image:Lethbridge 101021RFP.jpg|200px|right|Easy visual screening of transformed <i>Escherichia coli</i> cells using the Red/White 3-antibiotic assembly method]] | [[image:Lethbridge 101021RFP.jpg|200px|right|Easy visual screening of transformed <i>Escherichia coli</i> cells using the Red/White 3-antibiotic assembly method]] | ||
This method is a variant of the 3-Antibiotic Assembly Method that has been developed by Ginkgo Bioworks and New England Biolabs.<br> | This method is a variant of the 3-Antibiotic Assembly Method that has been developed by Ginkgo Bioworks and New England Biolabs.<br> | ||
| - | *In the 3-Antibiotic Assembly Method, the destination backbone (< | + | *In the 3-Antibiotic Assembly Method, the destination backbone (<html><a href="http://partsregistry.org/wiki/index.php/Part:pSB1A3" target="new"><font color="#00DC00"> pSB1A3</font></a></html>, <html><a href="http://partsregistry.org/wiki/index.php/Part:pSB1C3" target="new"><font color="#00DC00"> pSB1C3</font></a></html>, <html><a href="http://partsregistry.org/wiki/index.php/Part:pSB1K3" target="new"><font color="#00DC00"> pSB1K3</font></a></html>, or <html><a href="http://partsregistry.org/wiki/index.php/Part:pSB1T3" target="new"><font color="#00DC00"> pSB1T3</font></a></html>) is amplified via PCR, using DNA received in the 2010 Distribution. <br> |
We had difficulty generating a large quantity of plasmid backbone in this manner, and as a result were unable to assemble biobricks. <br><br> | We had difficulty generating a large quantity of plasmid backbone in this manner, and as a result were unable to assemble biobricks. <br><br> | ||
| - | As an alternative, we exploited the ability of part < | + | As an alternative, we exploited the ability of part <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_J04450" target="new"><font color="#00DC00"> BBa_J04450</font></a></html> (expressing red fluorescent protein - RFP) to create in its host cell a very strong red color following incubation; this has proven a useful and successful variant of the 3-Antibiotic Assembly Method.<br><br> |
We retained the selection advantage of the 3-Antibiotic Assembly Method (associated with having a destination plasmid containing a different antibiotic resistance than the upstream and downstream plasmid) as the expressing RFP biobrick is available in a wide variety of BBF plasmids. <br><br> | We retained the selection advantage of the 3-Antibiotic Assembly Method (associated with having a destination plasmid containing a different antibiotic resistance than the upstream and downstream plasmid) as the expressing RFP biobrick is available in a wide variety of BBF plasmids. <br><br> | ||
With this variation, we can not only visually screen our colonies for the presence of our expected assembly product (as indicated by a lack of red fluorescence), we also have a built-in negative control in that a lack of assembly product will result in a re-ligation of the expressing RFP biobrick into the destination backbone, producing red colonies.<br> | With this variation, we can not only visually screen our colonies for the presence of our expected assembly product (as indicated by a lack of red fluorescence), we also have a built-in negative control in that a lack of assembly product will result in a re-ligation of the expressing RFP biobrick into the destination backbone, producing red colonies.<br> | ||
| Line 183: | Line 183: | ||
Following each restriction digest, all samples were subjected to heating at 80<sup>o</sup>C for 20 minutes to irreversibly denature the restriction endonucleases.<br><br> | Following each restriction digest, all samples were subjected to heating at 80<sup>o</sup>C for 20 minutes to irreversibly denature the restriction endonucleases.<br><br> | ||
† Typical concentrations obtained from our minipreps are approximately 50ng/µL, therefore adding 5µL of plasmid DNA gives the restriction reaction a concentration of 10ng/µL, as recommended in the NEB BioBrick Assembly Kit Literature. <br> | † Typical concentrations obtained from our minipreps are approximately 50ng/µL, therefore adding 5µL of plasmid DNA gives the restriction reaction a concentration of 10ng/µL, as recommended in the NEB BioBrick Assembly Kit Literature. <br> | ||
| - | ‡ Destination plasmid is either pSB1A3, pSB1C3, pSB1K3, or pSB1T3, containing part < | + | ‡ Destination plasmid is either pSB1A3, pSB1C3, pSB1K3, or pSB1T3, containing part <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_J04450" target="new"><font color="#00DC00">BBa_J04450</font></a></html> which is the expressing red fluorescent protein. |
===<font color="white">Step 2 - Ligation=== | ===<font color="white">Step 2 - Ligation=== | ||
 "
"