Team:Lethbridge/Results
From 2010.igem.org
Liszabruder (Talk | contribs) |
Liszabruder (Talk | contribs) |
||
| Line 130: | Line 130: | ||
=<font color="white">Compartmentalization Parts= | =<font color="white">Compartmentalization Parts= | ||
| - | One of the sub-projects for the bioremediation of the tailings ponds is to create synthetic <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Compartamentalization"><font color=" | + | One of the sub-projects for the bioremediation of the tailings ponds is to create synthetic <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Compartamentalization"><font color="#00DC00"> microcompartments</font></a></html> that we can then use to isolate various pathway within an <i>Escherichia coli</i> cell. To do this we need to have a microcompartment as well as a means to characterize the compartment so that the system can be optimized. Here are the experiments we have performed so far towards the characterization of the microcomparments. |
<br><br> | <br><br> | ||
==<font color="white">Placement of Oligoarginine Tail on Proteins</font>== | ==<font color="white">Placement of Oligoarginine Tail on Proteins</font>== | ||
===<font color="white">Characterized Parts</font>=== | ===<font color="white">Characterized Parts</font>=== | ||
<hr> | <hr> | ||
| - | <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K249004" target="new"><font color=" | + | <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K249004" target="new"><font color="#00DC00" size="+1">BBa_K249004</font></a></html> |
<br><br> | <br><br> | ||
| - | <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K249005"target="new"><font color=" | + | <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K249005"target="new"><font color="#00DC00" size="+1">BBa_K249005</font></a></html> |
<br><br> | <br><br> | ||
| - | <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K331030"target="new"><font color=" | + | <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K331030"target="new"><font color="#00DC00" size="+1">BBa_K331030</font></a></html> |
<br><br> | <br><br> | ||
| - | <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K331031"target="new"><font color=" | + | <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K331031"target="new"><font color="#00DC00" size="+1">BBa_K331031</font></a></html> |
<br><br> | <br><br> | ||
===<font color="white">Hypothesis</font>=== | ===<font color="white">Hypothesis</font>=== | ||
| Line 149: | Line 149: | ||
===<font color="white">Introduction</font>=== | ===<font color="white">Introduction</font>=== | ||
<hr> | <hr> | ||
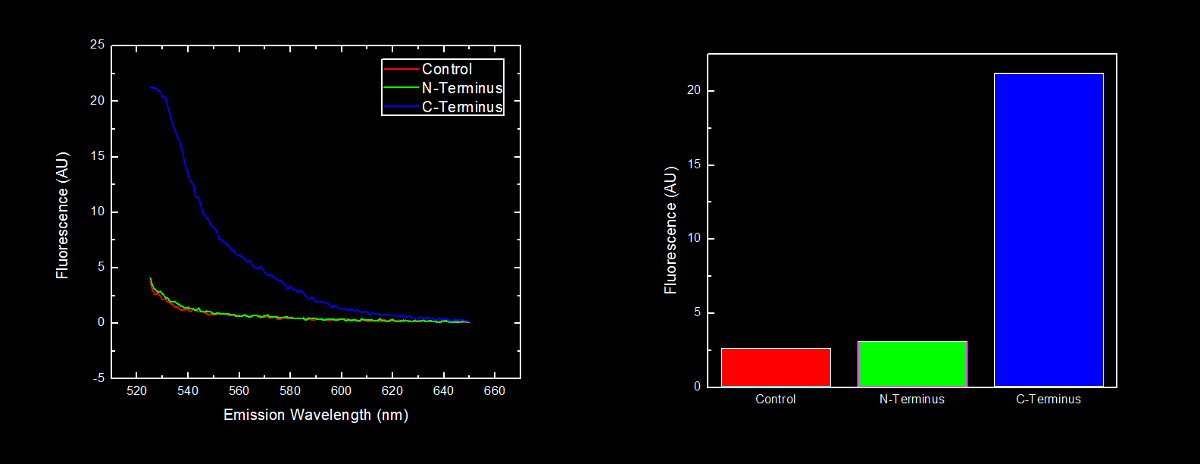
| - | The long term goal of our team is to utilize an oligoarginine tail to specifically target enzymes into a microcompartment composed of modified lumazine synthase subunits. While conducting background research on the project, we came upon data originally reported by Bachmair <i>et al.</i><sup>1</sup> suggesting that the identity of the amino acid at the N-terminus of a protein is related to its half-life, and mostly notably, that arginine residues at the are destabilizing. This data suggests that by placing an arginine at the N-terminus of a protein to be targeted into a <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Compartamentalization"><font color=" | + | The long term goal of our team is to utilize an oligoarginine tail to specifically target enzymes into a microcompartment composed of modified lumazine synthase subunits. While conducting background research on the project, we came upon data originally reported by Bachmair <i>et al.</i><sup>1</sup> suggesting that the identity of the amino acid at the N-terminus of a protein is related to its half-life, and mostly notably, that arginine residues at the are destabilizing. This data suggests that by placing an arginine at the N-terminus of a protein to be targeted into a <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Compartamentalization"><font color="#00DC00"> lumazine synthase microcompartment</font></a></html> would cause degradation of our protein before it can be moved into the microcompartment. |
<br><br> | <br><br> | ||
We chose to investigate the how the placement of an oligoarginine sequence affects the stability of the protein to which it is fused. | We chose to investigate the how the placement of an oligoarginine sequence affects the stability of the protein to which it is fused. | ||
| Line 156: | Line 156: | ||
===<font color="white">Method</font>=== | ===<font color="white">Method</font>=== | ||
<hr> | <hr> | ||
| - | In order to further characterize the C-terminal and N-terminal oligoarginine tag (BioBricks <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K249005" target="new"><font color=" | + | In order to further characterize the C-terminal and N-terminal oligoarginine tag (BioBricks <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K249005" target="new"><font color="#00DC00">BBa_K249005</font></a></html> and <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K249004" target="new"><font color="#00DC00">BBa_K249004</font></a></html> respectively) and investigate the effect their placement on protein stability, yellow fluorescent proteins (YFP) with the oligoarginine fused to either the C-terminus (<html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K331023" target="new"><font color="#00DC00">BBa_K331023</font></a></html>) or N-terminus (<html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K331022" target="new"><font color="#00DC00">BBa_K331022</font></a></html>) (and preceded by a ribosomal binding site – <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_B0034" target="new"><font color="#00DC00">BBa_B0034</font></a></html>) were synthesized. We used our <html><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Protocols#Assembly_of_BioBricks_using_the_Red.2FWhite_3-Antibiotic_Assembly_Method"><font color="#00DC00"> Red/White 3-Antibiotic assembly method</font></a></html> to add a tetracycline repressible promoter (<html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_R0010" target="new"><font color="#00DC00">BBa_R0010</font></a></html>) for constitutive expression of the fusion protein. This addition generated BioBricks <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K331031" target="new"><font color="#00DC00">BBa_K331031</font></a></html> and <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_K331030" target="new"><font color="#00DC00">BBa_K331030</font></a></html> for the C-terminal tagged and N-terminal tagged YFP respectively. |
<br><br> | <br><br> | ||
The BioBrick containing plasmid was transformed into <i>Escherichia coli</i> DH5α cells. These cells were grown to an OD<sub>600</sub> of approximately 0.7, and diluted 1:10 with MilliQ H2O immediately prior to analysis by fluorescent spectroscopy. | The BioBrick containing plasmid was transformed into <i>Escherichia coli</i> DH5α cells. These cells were grown to an OD<sub>600</sub> of approximately 0.7, and diluted 1:10 with MilliQ H2O immediately prior to analysis by fluorescent spectroscopy. | ||
 "
"