Team:Heidelberg/Modeling
From 2010.igem.org
AlejandroHD (Talk | contribs) |
AlejandroHD (Talk | contribs) |
||
| Line 19: | Line 19: | ||
The seed of the miRNA is usually defined as the region centered in the nucleotides 2-7 in the 5’ end of the miRNA, and it usually requires extensive pairing. For the sake of simplicity, we extended slightly the term seed to include the nucleotides 1-8. | The seed of the miRNA is usually defined as the region centered in the nucleotides 2-7 in the 5’ end of the miRNA, and it usually requires extensive pairing. For the sake of simplicity, we extended slightly the term seed to include the nucleotides 1-8. | ||
In miBS designer, the user can choose between several types of seed to create their binding site (ordered by increasing efficacy): | In miBS designer, the user can choose between several types of seed to create their binding site (ordered by increasing efficacy): | ||
| - | - 6mer (abundance 21.5%): only the nucleotides 2-7 of the miRNA match with the mRNA. | + | |
| - | - 7merA1 (abundance 15.1%): the nucleotides 2-7 match with the mRNA, and there is an adenine in position 1. | + | -6mer (abundance 21.5%): only the nucleotides 2-7 of the miRNA match with the mRNA. |
| - | - 7merm8 (abundance 25%): the nucleotides 2-8 match with the mRNA. | + | |
| - | - 8mer (abundance 19.8%): the nucleotides 2-8 match with the mRNA and there is an adenine in position 1. | + | -7merA1 (abundance 15.1%): the nucleotides 2-7 match with the mRNA, and there is an adenine in position 1. |
| - | - Apart from any of these options, the user can decide to create a customized seed with a mismatch included. | + | |
| + | -7merm8 (abundance 25%): the nucleotides 2-8 match with the mRNA. | ||
| + | |||
| + | -8mer (abundance 19.8%): the nucleotides 2-8 match with the mRNA and there is an adenine in position 1. | ||
| + | |||
| + | -Apart from any of these options, the user can decide to create a customized seed with a mismatch included. | ||
*The percentages of abundance are calculated among conserved mammalian sites for a highly conserved miRNA (Friedman et al. 2008) | *The percentages of abundance are calculated among conserved mammalian sites for a highly conserved miRNA (Friedman et al. 2008) | ||
| Line 40: | Line 45: | ||
| - | + | ==Neural Network Model== | |
| - | + | ===Neural Network theory=== | |
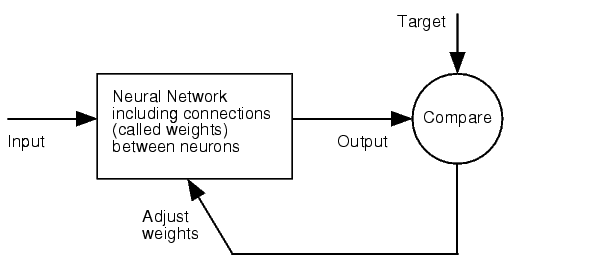
Artificial Neural Network usually called (NN), is a computational model that is inspired by the biological nervous system. The network is composed of simple elements called artificial neurons that are interconnected and operate in parallel. In most cases the NN is an adaptive system that can change its structure depending on the internal or(and?) external information that flows into the network during the learning process. The NN can be trained to perform a particular function by adjusting the values of the connection, called weights, between the artificial neurons. Neural Networks have been employed to perform complex functions in various fields, including pattern recognition, identification, classification, speech, vision, and control systems. | Artificial Neural Network usually called (NN), is a computational model that is inspired by the biological nervous system. The network is composed of simple elements called artificial neurons that are interconnected and operate in parallel. In most cases the NN is an adaptive system that can change its structure depending on the internal or(and?) external information that flows into the network during the learning process. The NN can be trained to perform a particular function by adjusting the values of the connection, called weights, between the artificial neurons. Neural Networks have been employed to perform complex functions in various fields, including pattern recognition, identification, classification, speech, vision, and control systems. | ||
| Line 49: | Line 54: | ||
Figure 1: Training of a Neural Network. | Figure 1: Training of a Neural Network. | ||
| - | + | ===Model description=== | |
The NN model has been created with the MATLAB NN-toolbox. The input/target pairs used to train the network comprise experimental and literature data (Bartel et al. 2007). The experimental data were obtained by measuring via luciferase assay the strength of knockdown due to the interaction between the shRNA and the binding site situated on the 3’UTR of luciferase gene. Nearly 30 different rational designed binding sites were tested and the respective knockdown strength calculated with the following formula->(formula anyone???). | The NN model has been created with the MATLAB NN-toolbox. The input/target pairs used to train the network comprise experimental and literature data (Bartel et al. 2007). The experimental data were obtained by measuring via luciferase assay the strength of knockdown due to the interaction between the shRNA and the binding site situated on the 3’UTR of luciferase gene. Nearly 30 different rational designed binding sites were tested and the respective knockdown strength calculated with the following formula->(formula anyone???). | ||
| - | + | ====Input/target pairs==== | |
Each input was represented by a four elements vector. Each element corresponded to a score value related to a specific feature of the binding site. The four features used to describe the binding site were: seed type, the 3’pairing contribution the AU-content and the number of binding site. The input/target pair represented the relationship between a particular binding site and the related percentage of knockdown. | Each input was represented by a four elements vector. Each element corresponded to a score value related to a specific feature of the binding site. The four features used to describe the binding site were: seed type, the 3’pairing contribution the AU-content and the number of binding site. The input/target pair represented the relationship between a particular binding site and the related percentage of knockdown. | ||
Once the network was trained than it was used to predict percentages of knockdown given certain inputs. The predictions were then validated experimentally. | Once the network was trained than it was used to predict percentages of knockdown given certain inputs. The predictions were then validated experimentally. | ||
| - | + | ===Characteristic of the Network=== | |
| - | + | ===Results=== | |
| - | + | ==Fuzzy Inference Model== | |
[[#shRNA_binding_sites|shRNA binding sites]] | [[#shRNA_binding_sites|shRNA binding sites]] | ||
Revision as of 05:22, 26 October 2010

|
|
||||||||||||
 "
"