Team:Newcastle/End of crack & signalling system
From 2010.igem.org
(→End of crack cell-signalling system) |
Swoodhouse (Talk | contribs) (→Subtilin Production part) |
||
| (36 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
=End of crack cell-signalling system= | =End of crack cell-signalling system= | ||
| - | Subtilin | + | Subtilin belongs to a class of peptide antibiotics, called lantibiotic that contain polycyclic thioether amino acids as well as the unsaturated amino acids dehydroalanine and 2-aminoisobutyric acid. It is produced by ''Bacillus subtilis'' ATCC 6633, which is effective against a wide range of Gram positive bacteria including pathogenic bacteria such as propionibacteria, staphylococci, clostridia, enterococci and streptococci. |
| + | Subtilin also doubles up as a quorum sensing molecule, which we can use to trigger calcium carbonate precipitation and filament cell formation once the bacteria have reached a sufficient density inside a crack. We developed a subtilin immunity part and a subtilin production part to add to the already-existing [http://partsregistry.org/Part:BBa_K104001 subtilin sensing part on the parts registry]. | ||
| + | [[Image:Newcastle subtilin diagram.png|700px]] | ||
| - | + | '''Figure 1:''' Schematic diagram of subtilin cell signalling system in ''B. subtilis'' 168 | |
| - | |||
| - | |||
| - | == | + | ==Subtilin Production part== |
| - | + | [[Image:Subtilin_production.png|700px]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The subtilin production Biobrick consists of the ''spaBCTS'' gene cluster under an oxygen limitation sensitive promoter. In low oxygen conditions, similar to conditions at the end of a crack with high cell density, ''spaS'' will be expressed. The immature SpaS will then have to be modified and transported out of the cell by SpaBCT, an ABC transporter. | |
| - | + | We will place this part in front of the existing [http://partsregistry.org/Part:BBa_K104001 subtilin-sensor part], allowing the subtilin signal to propagate from the end of the crack throughout the cell population. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | = | + | This is [http://partsregistry.org/wiki/index.php?title=Part:BBa_K302021 BBa_K302021 on the parts registry]. |
| - | + | ==Subtilin Immunity part== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[Image: | + | [[Image:subtilin_immunity.png|700px]] |
| - | + | The subtilin immunity BioBrick consists of the ''spaIFEG'' gene cluster under a constitutive promoter, Pveg. Immunity is based on two systems. SpaI is a lipoprotein that will bind to and interacts with subtilin at the cell surface. SpaFEG forms an ABC transporter homologue that will expel subtilin from the cell membrane into the extra cellular matrix. | |
| - | = | + | This is [http://partsregistry.org/wiki/index.php?title=Part:BBa_K302018 BBa_K302018 on the parts registry]. |
==Cloning Strategy== | ==Cloning Strategy== | ||
| Line 51: | Line 35: | ||
[[Media:Newcastle CloningStrategy_BioBrick_Subtilin_Immunity.pdf|Subtilin Immunity]] | [[Media:Newcastle CloningStrategy_BioBrick_Subtilin_Immunity.pdf|Subtilin Immunity]] | ||
| - | + | ==Testing and Characterisation== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
====Selection for integration==== | ====Selection for integration==== | ||
| - | To select for integration of the plasmid into the chromosome, ''B. subtilis'' will be tested for the ability to hydrolyse starch. Integration of the BioBrick will be done by homologous recombination at the amyE gene, therefore destroying | + | To select for integration of the plasmid into the chromosome, ''B. subtilis'' will be tested for the ability to hydrolyse starch. Integration of the BioBrick will be done by homologous recombination at the ''amyE'' gene, therefore destroying the ability to express amylase. Colonies that do not contain the integrated BioBrick will be able to hydrolyse starch, therefore forming a white halo around the colony as iodine interacts with starch to form blue colour. |
| - | + | ====Isolation of subtilin from ''B. subtilis'' ATCC 6633==== | |
| - | ====Isolation of subtilin from ''B. subtilis'' | + | {| |
| - | + | |1. ''B. subtilis'' ATCC6633 were grown in 250 ml flasks containing 100 ml of BHI broth and incubate for 48 h at 32°C in shaker at 125 cycles/min. | |
| - | 1. ''B. subtilis'' ATCC6633 were grown in 250 ml flasks containing 100 ml of BHI broth and incubate for 48 h at 32°C in shaker at 125 cycles/min. | + | |- |
| - | 2. Culture media were centrifuged at 10000g for 15 min and the supernatants were filtered through 0.22 | + | |2. Culture media were centrifuged at 10000g for 15 min and the supernatants were filtered through 0.22 um membrane. |
| - | 3. The pH of the filtrates has to be between 7.0 to 8.0 pH. | + | |- |
| - | 4. Store the filtrates at -20°C for further use. | + | |3. The pH of the filtrates has to be between 7.0 to 8.0 pH. |
| + | |- | ||
| + | |4. Store the filtrates at -20°C for further use. | ||
| + | |} | ||
===Characterisation of BioBrick=== | ===Characterisation of BioBrick=== | ||
| - | + | {| | |
| - | 1. Make up 4 LB agar plates and inoculate with the following organism: | + | |1. Make up 4 LB agar plates and inoculate with the following organism:Negative Control –'' B. subtilis'' 168, Positive Control – ''B. subtilis'' ATCC6633 and Test (Duplicates) – ''B. subtilis'' 168 containing subtilin immunity BioBrick. |
| - | + | |- | |
| - | + | |2. Apply 20 ul of the cell free filtrate containing subtilin onto cellulose disks (6 mm) and place onto the LB agar plates. | |
| - | + | |- | |
| - | 2. Apply 20 | + | |3. Plates were incubated for 24 h at 37 °C. |
| - | 3. Plates were incubated for 24 h at 37 °C. | + | |- |
| - | 4. Zone of inhibition were then measured. | + | |4. Zone of inhibition were then measured. |
| + | |} | ||
====Expected results==== | ====Expected results==== | ||
| Line 107: | Line 77: | ||
A.S. Motta and A. Brandelli. (2002) Characterization of an antibacterial peptide produced by Brevibacterium linens. Journal of Applied Microbiology 92, 63-70. | A.S. Motta and A. Brandelli. (2002) Characterization of an antibacterial peptide produced by Brevibacterium linens. Journal of Applied Microbiology 92, 63-70. | ||
| - | |||
A.S. Motta, F. Cladera-Olivera and A. Brandelli (2004) Screen for antimicrobial activity among bacteria isolated from the amazon basin. Brazilian Journal of Microbiology 35, 307-310. | A.S. Motta, F. Cladera-Olivera and A. Brandelli (2004) Screen for antimicrobial activity among bacteria isolated from the amazon basin. Brazilian Journal of Microbiology 35, 307-310. | ||
Latest revision as of 00:56, 26 October 2010

| |||||||||||||
| |||||||||||||
Contents |
End of crack cell-signalling system
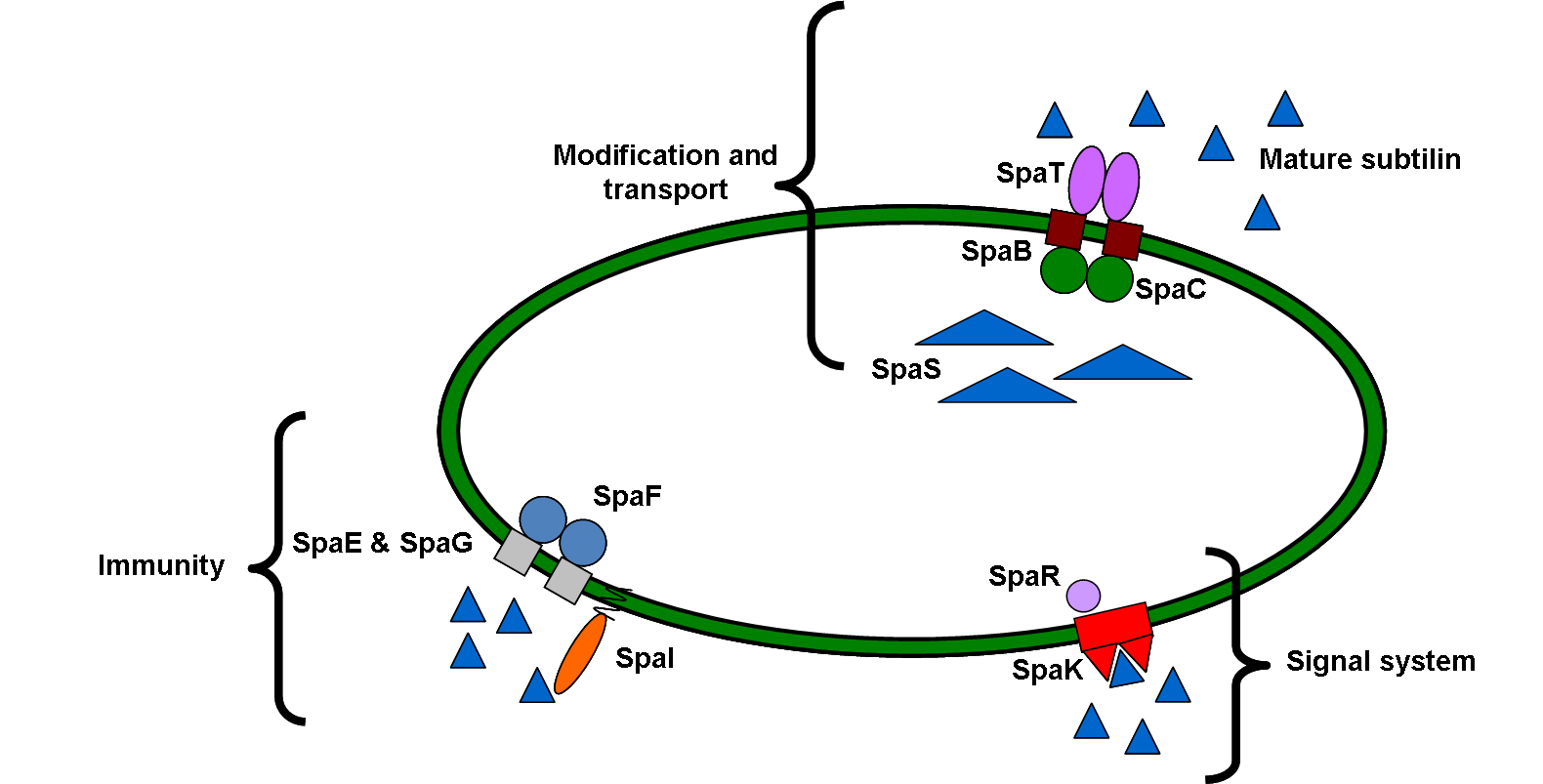
Subtilin belongs to a class of peptide antibiotics, called lantibiotic that contain polycyclic thioether amino acids as well as the unsaturated amino acids dehydroalanine and 2-aminoisobutyric acid. It is produced by Bacillus subtilis ATCC 6633, which is effective against a wide range of Gram positive bacteria including pathogenic bacteria such as propionibacteria, staphylococci, clostridia, enterococci and streptococci.
Subtilin also doubles up as a quorum sensing molecule, which we can use to trigger calcium carbonate precipitation and filament cell formation once the bacteria have reached a sufficient density inside a crack. We developed a subtilin immunity part and a subtilin production part to add to the already-existing [http://partsregistry.org/Part:BBa_K104001 subtilin sensing part on the parts registry].
Figure 1: Schematic diagram of subtilin cell signalling system in B. subtilis 168
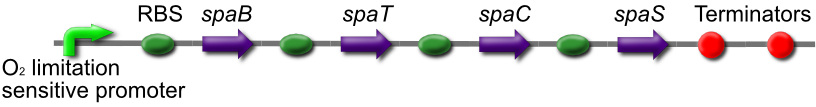
Subtilin Production part
The subtilin production Biobrick consists of the spaBCTS gene cluster under an oxygen limitation sensitive promoter. In low oxygen conditions, similar to conditions at the end of a crack with high cell density, spaS will be expressed. The immature SpaS will then have to be modified and transported out of the cell by SpaBCT, an ABC transporter.
We will place this part in front of the existing [http://partsregistry.org/Part:BBa_K104001 subtilin-sensor part], allowing the subtilin signal to propagate from the end of the crack throughout the cell population.
This is [http://partsregistry.org/wiki/index.php?title=Part:BBa_K302021 BBa_K302021 on the parts registry].
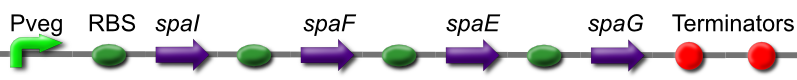
Subtilin Immunity part
The subtilin immunity BioBrick consists of the spaIFEG gene cluster under a constitutive promoter, Pveg. Immunity is based on two systems. SpaI is a lipoprotein that will bind to and interacts with subtilin at the cell surface. SpaFEG forms an ABC transporter homologue that will expel subtilin from the cell membrane into the extra cellular matrix.
This is [http://partsregistry.org/wiki/index.php?title=Part:BBa_K302018 BBa_K302018 on the parts registry].
Cloning Strategy
Testing and Characterisation
Selection for integration
To select for integration of the plasmid into the chromosome, B. subtilis will be tested for the ability to hydrolyse starch. Integration of the BioBrick will be done by homologous recombination at the amyE gene, therefore destroying the ability to express amylase. Colonies that do not contain the integrated BioBrick will be able to hydrolyse starch, therefore forming a white halo around the colony as iodine interacts with starch to form blue colour.
Isolation of subtilin from B. subtilis ATCC 6633
| 1. B. subtilis ATCC6633 were grown in 250 ml flasks containing 100 ml of BHI broth and incubate for 48 h at 32°C in shaker at 125 cycles/min. |
| 2. Culture media were centrifuged at 10000g for 15 min and the supernatants were filtered through 0.22 um membrane. |
| 3. The pH of the filtrates has to be between 7.0 to 8.0 pH. |
| 4. Store the filtrates at -20°C for further use. |
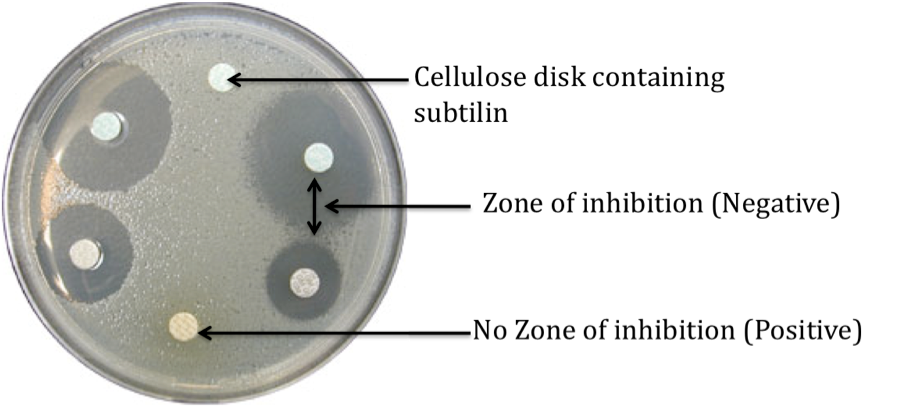
Characterisation of BioBrick
| 1. Make up 4 LB agar plates and inoculate with the following organism:Negative Control – B. subtilis 168, Positive Control – B. subtilis ATCC6633 and Test (Duplicates) – B. subtilis 168 containing subtilin immunity BioBrick. |
| 2. Apply 20 ul of the cell free filtrate containing subtilin onto cellulose disks (6 mm) and place onto the LB agar plates. |
| 3. Plates were incubated for 24 h at 37 °C. |
| 4. Zone of inhibition were then measured. |
Expected results
| 1. Negative control – Zone of inhibition to be observed |
| 2. Positive control – Zone of inhibition not observed |
| 3. Test (Duplicates) – Zone of inhibition not observed or a smaller zone of inhibition as compared to the negative control |
References
A.S. Motta and A. Brandelli. (2002) Characterization of an antibacterial peptide produced by Brevibacterium linens. Journal of Applied Microbiology 92, 63-70.
A.S. Motta, F. Cladera-Olivera and A. Brandelli (2004) Screen for antimicrobial activity among bacteria isolated from the amazon basin. Brazilian Journal of Microbiology 35, 307-310.
 
|
 "
"