Team:Northwestern/Project/Modeling
From 2010.igem.org
(Difference between revisions)
| Line 162: | Line 162: | ||
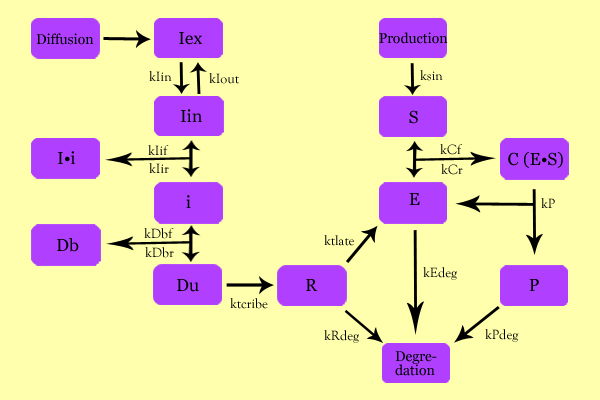
In terms of our iGEM project, this model was employed to explore the effect of IPTG concentration and diffusion, lacI concentration (determined by the combination part of constitutive promoter, ribosome binding site, lacI gene, double terminator, lac promoter/operon), and the ribosome binding site on the concentrations of all species involved - described in the following sections - and especially on Chitin Synthase and Chitin concentration and the corresponding rates. | In terms of our iGEM project, this model was employed to explore the effect of IPTG concentration and diffusion, lacI concentration (determined by the combination part of constitutive promoter, ribosome binding site, lacI gene, double terminator, lac promoter/operon), and the ribosome binding site on the concentrations of all species involved - described in the following sections - and especially on Chitin Synthase and Chitin concentration and the corresponding rates. | ||
| + | |||
=='''Modeling'''== | =='''Modeling'''== | ||
| Line 170: | Line 171: | ||
[[Image:SuperModeling.jpg|600px|center]] | [[Image:SuperModeling.jpg|600px|center]] | ||
| + | |||
==='''Variables'''=== | ==='''Variables'''=== | ||
| Line 184: | Line 186: | ||
* C: Enzyme Substrate Complex (CHS3-(N-Acetyl-Glucosamine)-Chitin or (NAG)n Complex) | * C: Enzyme Substrate Complex (CHS3-(N-Acetyl-Glucosamine)-Chitin or (NAG)n Complex) | ||
* P: Protein Product (Chitin or (NAG)n+1) | * P: Protein Product (Chitin or (NAG)n+1) | ||
| + | |||
==='''Equations'''=== | ==='''Equations'''=== | ||
| Line 199: | Line 202: | ||
* dCC = kCf*E*Si - kCr*CC - kP*CC; | * dCC = kCf*E*Si - kCr*CC - kP*CC; | ||
* dP = kP*CC - kPdeg*P; | * dP = kP*CC - kPdeg*P; | ||
| + | |||
==='''IPTG Diffusion'''=== | ==='''IPTG Diffusion'''=== | ||
First, Fick's Law of Diffusion was modeled through MATLAB. The diffusion constant used was 220um^2/s.[4] | First, Fick's Law of Diffusion was modeled through MATLAB. The diffusion constant used was 220um^2/s.[4] | ||
| - | |||
[[Image:loopydoops.gif]] | [[Image:loopydoops.gif]] | ||
| - | |||
IPTG was sprayed at the top of the colony, which then diffuses as according to Fick's law. The spatial local concentration will then differentially induce downstream processes. | IPTG was sprayed at the top of the colony, which then diffuses as according to Fick's law. The spatial local concentration will then differentially induce downstream processes. | ||
| + | |||
==='''Semi-Empirical Determination'''=== | ==='''Semi-Empirical Determination'''=== | ||
| - | + | Status: Under Development | |
| + | |||
| + | The initial plan was to use lacI-constitutive expression / lac-operon (CP-LacpI) part with Green Fluorescent Protein to acquire empirical data. | ||
| + | |||
| + | By testing various combinations of CP/LacpI, RBS, and IPTG concentrations with GFP, one could acquire fluorescence data over time using a plate reader, and use that data to fit the current model, where instead of producing Chitin Synthase, the protein product would be GFP, the concentration of which could be measured as fluorescence. | ||
| + | |||
| + | However, at the time of the wiki-freeze, data acquisition is incomplete. | ||
| - | |||
==='''Fitted Model'''=== | ==='''Fitted Model'''=== | ||
| Line 221: | Line 229: | ||
This model was fitted with empirical data using cp-lacpi-gfp to estimate the rate constant, and therefore the effects of varying cp, lacpi, and rbs on enzyme and final product production. | This model was fitted with empirical data using cp-lacpi-gfp to estimate the rate constant, and therefore the effects of varying cp, lacpi, and rbs on enzyme and final product production. | ||
| + | |||
=='''References'''== | =='''References'''== | ||
Revision as of 22:52, 25 October 2010
| Home | Brainstorm | Team | Acknowledgements | Project | Human Practices | Parts | Notebook | Calendar | Protocol | Safety | Links | References | Media | Contact |
|---|
 "
"