Project/PlcR Promoter/
From 2010.igem.org
(New page: == PlcR Promoter == <br /> === Background === <br /> === Sequence Development and Annotation === <br /> === Testing and Validation === <br /> === References === <br />) |
|||
| (10 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | == PlcR Promoter == | + | {{Template:WITS-South_Africa_Main_Menu_19_07}} |
| - | + | {{Template:WITS-South_Africa_Main_Menu_0910}} | |
| + | |||
| + | == PlcR Promoter - [http://partsregistry.org/Part:BBa_K354003 BBa_K354003] == | ||
=== Background === | === Background === | ||
| - | <br /> | + | Derived from Bacillis thuringiensis, the plcR promoter regulates a host of genes required for the initiation of virulence in response to specific environmental cues [1]. The plcR promoter has two identifiable binding sequences for both the PlcR-PapR fusion peptide as well as SpoOA repressor protein. Binding of SpoOA to PlcR promoter blocks the PlcR-PapR fusion peptide binding sequence, thus preventing the adhesion of RNA polymerase and transcription. When PlcR-PapR is present, SpoOA is prevented from binding and thus the polymerase-DNA interaction happens freely and transcription of downstream genes is positively regulated. Ten bases upstream of the start codon lies the consensus sequence for the Sigma-A factor of RNA polymerase II. <br /> |
| + | |||
=== Sequence Development and Annotation === | === Sequence Development and Annotation === | ||
| + | In the same way that we assembled the Lac/Ara-1 promoter from primer-primer adhesion and their subsequent extension, the PlcR promoter was assembled in the same fashion. See the [https://2010.igem.org/Project/Lac1/AraC_Promoter/ Lac/Ara -1 Promoter] for the full assembly details. | ||
| + | |||
| + | <div class="heading"> | ||
| + | '''Promoter Assembly''' | ||
| + | </div> | ||
| + | The forward primer used in assembly is: <br /> | ||
| + | [[Image:Plcr_forward_promoter.JPG]] <br /> | ||
| + | The sequence in red is the EcoR1 restriction site with that in blue being the Xba1 restriction site. The region in purple are the bases that the forward and reverse primers overlap with one another. <br /> <br /> | ||
| + | |||
| + | The reverse primer used in assembly is: <br /> | ||
| + | [[Image:Plcr_reverse_promoter.JPG]] <br /> | ||
| + | The sequence in green is a Pst1 restriction site and that in orange is the Spe1 restriction site. The region in purple is the sequence of shared overlap with the forward primer. As can be seen, there have been randomised nucleotides introduced so as to create a library of varying promoters. This was performed so as to increase the stochastic nature associated with the promoter efficacy. According to the IUPAC nomenclature, Y indicates either a C or a T while W indicates either an A or a T. <br /> <br /> | ||
| + | |||
| + | The entire PlcR promoter sequence is: <br /> | ||
| + | [[Image:Plcr_promoter_full.JPG]] <br /> | ||
| + | As can be seen, the 'g' in red resulted from the insertion of a C and the 't' in green from the insertion of an A. | ||
<br /> | <br /> | ||
| + | |||
| + | <div class="heading"> | ||
| + | '''The Package''' | ||
| + | </div> | ||
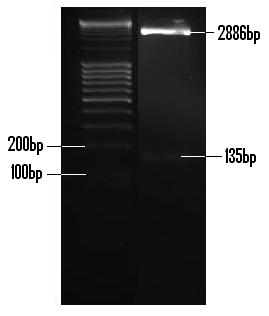
| + | Once synthesized via primer extension, the PlcR promoter was ligated into the PTZ plasmid, transformed and then extracted in an appreciable amount. The insert was digested with EcoR1 and Pst1, gel extracted and purified. The PlcR promoter insert (135bp) and linearised pTZ57R/T (2886bp) are visible. <br /> | ||
| + | [[Image:PlcR_Promoter_digested.JPG]] <br /> <br /> | ||
| + | |||
| + | The following confirmation of the successful ligation of the PlcR promoter into PSB1C3 is in a 2% (m/v) agarose gel. <br /> | ||
| + | [[Image:Plcr_confirmation_gel.JPG]] <br /> | ||
| + | The above gel indicates the PlcR promoter (135bp) and the linearised PSB1C3 plasmid (2072bp). The fragments were isolated by digestion with EcoR1 and Pst1. | ||
| + | |||
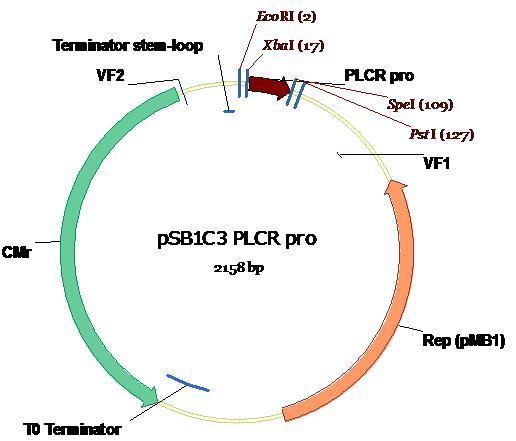
| + | The plasmid map, annotated with appropriate restriction sites and sizes depicts the PlcR promoter ligated into PSB1C3. <br /> | ||
| + | [[Image:PlcR_in_PSB1C3.JPG]] <br /> <br /> | ||
| + | |||
| + | |||
=== Testing and Validation === | === Testing and Validation === | ||
<br /> | <br /> | ||
=== References === | === References === | ||
| - | <br /> | + | [1] Lereclus D., Agaisse H., Gominet M., Salamitou S. & Sanchis V. (1996). Identification of a Bacillus thuringiensis Gene That Positively Regulates Transcription of the Phosphatidylinostitol-Specific Phospholipase C Gene at the Onset of the Stationary Phase. Journal of bacteriology '''178:'''2749-2756. <br /> |
Latest revision as of 20:31, 25 October 2010

Contents |
PlcR Promoter - [http://partsregistry.org/Part:BBa_K354003 BBa_K354003]
Background
Derived from Bacillis thuringiensis, the plcR promoter regulates a host of genes required for the initiation of virulence in response to specific environmental cues [1]. The plcR promoter has two identifiable binding sequences for both the PlcR-PapR fusion peptide as well as SpoOA repressor protein. Binding of SpoOA to PlcR promoter blocks the PlcR-PapR fusion peptide binding sequence, thus preventing the adhesion of RNA polymerase and transcription. When PlcR-PapR is present, SpoOA is prevented from binding and thus the polymerase-DNA interaction happens freely and transcription of downstream genes is positively regulated. Ten bases upstream of the start codon lies the consensus sequence for the Sigma-A factor of RNA polymerase II.
Sequence Development and Annotation
In the same way that we assembled the Lac/Ara-1 promoter from primer-primer adhesion and their subsequent extension, the PlcR promoter was assembled in the same fashion. See the Lac/Ara -1 Promoter for the full assembly details.
Promoter Assembly
The forward primer used in assembly is:
The sequence in red is the EcoR1 restriction site with that in blue being the Xba1 restriction site. The region in purple are the bases that the forward and reverse primers overlap with one another.
The reverse primer used in assembly is:
The sequence in green is a Pst1 restriction site and that in orange is the Spe1 restriction site. The region in purple is the sequence of shared overlap with the forward primer. As can be seen, there have been randomised nucleotides introduced so as to create a library of varying promoters. This was performed so as to increase the stochastic nature associated with the promoter efficacy. According to the IUPAC nomenclature, Y indicates either a C or a T while W indicates either an A or a T.
The entire PlcR promoter sequence is:
As can be seen, the 'g' in red resulted from the insertion of a C and the 't' in green from the insertion of an A.
The Package
Once synthesized via primer extension, the PlcR promoter was ligated into the PTZ plasmid, transformed and then extracted in an appreciable amount. The insert was digested with EcoR1 and Pst1, gel extracted and purified. The PlcR promoter insert (135bp) and linearised pTZ57R/T (2886bp) are visible.
The following confirmation of the successful ligation of the PlcR promoter into PSB1C3 is in a 2% (m/v) agarose gel.
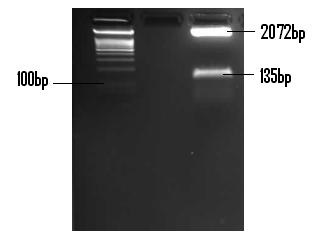
The above gel indicates the PlcR promoter (135bp) and the linearised PSB1C3 plasmid (2072bp). The fragments were isolated by digestion with EcoR1 and Pst1.
The plasmid map, annotated with appropriate restriction sites and sizes depicts the PlcR promoter ligated into PSB1C3.
Testing and Validation
References
[1] Lereclus D., Agaisse H., Gominet M., Salamitou S. & Sanchis V. (1996). Identification of a Bacillus thuringiensis Gene That Positively Regulates Transcription of the Phosphatidylinostitol-Specific Phospholipase C Gene at the Onset of the Stationary Phase. Journal of bacteriology 178:2749-2756.
 "
"