Project/Venus/
From 2010.igem.org
(New page: == Characterisation of the fliuorescent reporter protein - Venus == === BioBrick Background === <br /> === Sequence and Annotation === <br /> === Testing and verification === <br />) |
(→Characterisation of the fluorescent reporter protein - Venus - BBa_K354002) |
||
| (10 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | == Characterisation of the | + | {{Template:WITS-South_Africa_Main_Menu_19_07}} |
| + | {{Template:WITS-South_Africa_Main_Menu_0910}} | ||
| + | |||
| + | == Characterisation of the fluorescent reporter protein - Venus - [http://partsregistry.org/Part:BBa_K354002 BBa_K354002] == | ||
=== BioBrick Background === | === BioBrick Background === | ||
| - | <br /> | + | Depending on where you are on the spinning globe, Dear Reader, take a glance in the evening sky and find the brightest orb you can - Venus. <br /> <br /> |
| - | === Sequence and Annotation === | + | |
| - | <br /> | + | Derived from the yellow fluorescent protein (YFP), Venus is an enhanced version of the yellow-emitting fluorophore so commonly used as a reporter protein [1]. So as to gauge the rate of transcription, and hence rate of production of a string of genes in the same operon as Venus, the inclusion of this reporter allows our machine to be 'debugged' during testing. Chosen because of its fast folding and maturation time, Venus will produce a bright and transient a fluorescence when excited with the appropriate wavelength of incident light. For the exact excitation:emission values, see the BioBrick annotation page. |
| + | |||
| + | === Sequence Development and Annotation === | ||
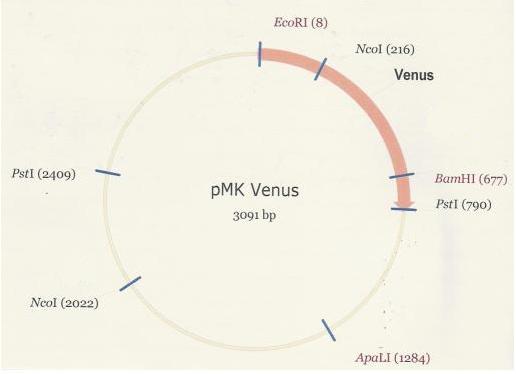
| + | As was the case in the assembly of the PlcR-PapR BioBrick, the synthesized Venus BioBrick was received in the pMK plasmid (below). | ||
| + | [[Image:PMK-Venus.JPG]] <br /> <br /> | ||
| + | |||
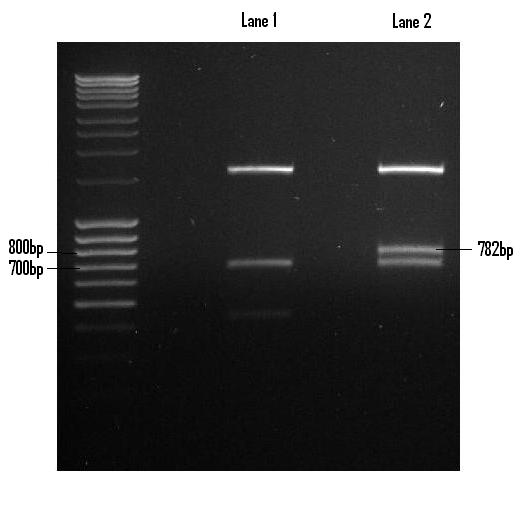
| + | Once digested with EcoR1 and Pst1, the Venus fragment was recovered via gel extraction and purified in preparation for its ligation into PSB1C3. Lane 2 houses the Venus digest with the excised fragment shown (782bp). <br /> | ||
| + | [[Image:Venus_digest.JPG]] <br /> <br /> | ||
| + | |||
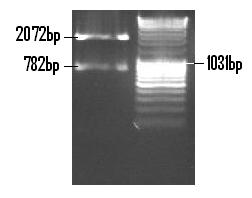
| + | The Venus fragment was ligated into the linearised form of PSB1C3 (digested with EcoR1 and Pst1) and the re-digested with EcoR1 and Pst1 so to confirm the successful ligation. The 782bp fragment confirms the ligation of Venus while the 2072bp fragment is indicative of the PSB1C3 backbone. <br /> | ||
| + | [[Image:Venus_in_PSB1C3.JPG]] <br /> <br /> | ||
| + | |||
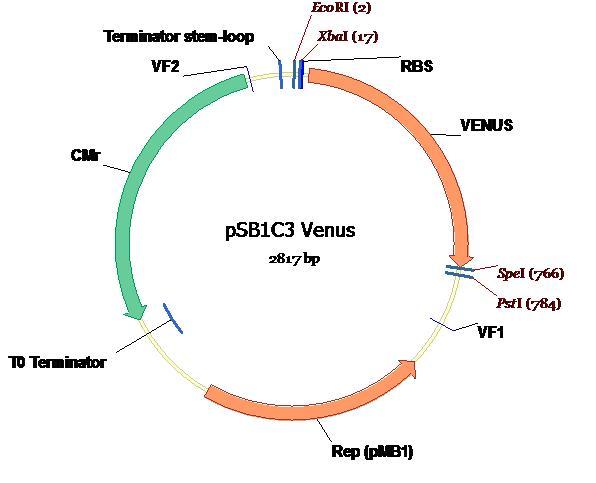
| + | The completed ligation of Venus into the PSB1C3 plasmid is depicted on the plasmid map (below) along with all annotations and landmark features [2]. <br /> | ||
| + | [[Image:Venus_plasmid_map.JPG]] <br /> | ||
| + | |||
| + | |||
| + | |||
=== Testing and verification === | === Testing and verification === | ||
| - | <br /> | + | As seen below, when driven by an appropriate promoter, Venus emits its characteristic fluorescence when excited by an appropriate wavelength of incident light. ''E. coli'' containing Venus can be distinguished from the background upon excitation. <br /> |
| + | [[Image:IGem_Venus_IPTG_T1hr_z0013_w0000.jpg]] <br /> | ||
| + | |||
| + | === References === | ||
| + | [1] Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K. & Miyawaki A. (2002), A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotechnology '''20:''' 87-90. <br /> | ||
| + | [2] Marco Weinberg, PhD. Antiviral Gene Therapy Research Unit, Department of Molecular Medicine and Haematology, University of the Witwatersrand Medical School, 7 York Rd. Parktown 2193 SOUTH AFRICA, marc.weinberg@wits.ac.za. | ||
Latest revision as of 20:12, 25 October 2010

Contents |
Characterisation of the fluorescent reporter protein - Venus - [http://partsregistry.org/Part:BBa_K354002 BBa_K354002]
BioBrick Background
Depending on where you are on the spinning globe, Dear Reader, take a glance in the evening sky and find the brightest orb you can - Venus.
Derived from the yellow fluorescent protein (YFP), Venus is an enhanced version of the yellow-emitting fluorophore so commonly used as a reporter protein [1]. So as to gauge the rate of transcription, and hence rate of production of a string of genes in the same operon as Venus, the inclusion of this reporter allows our machine to be 'debugged' during testing. Chosen because of its fast folding and maturation time, Venus will produce a bright and transient a fluorescence when excited with the appropriate wavelength of incident light. For the exact excitation:emission values, see the BioBrick annotation page.
Sequence Development and Annotation
As was the case in the assembly of the PlcR-PapR BioBrick, the synthesized Venus BioBrick was received in the pMK plasmid (below).
Once digested with EcoR1 and Pst1, the Venus fragment was recovered via gel extraction and purified in preparation for its ligation into PSB1C3. Lane 2 houses the Venus digest with the excised fragment shown (782bp).
The Venus fragment was ligated into the linearised form of PSB1C3 (digested with EcoR1 and Pst1) and the re-digested with EcoR1 and Pst1 so to confirm the successful ligation. The 782bp fragment confirms the ligation of Venus while the 2072bp fragment is indicative of the PSB1C3 backbone.
The completed ligation of Venus into the PSB1C3 plasmid is depicted on the plasmid map (below) along with all annotations and landmark features [2].
Testing and verification
As seen below, when driven by an appropriate promoter, Venus emits its characteristic fluorescence when excited by an appropriate wavelength of incident light. E. coli containing Venus can be distinguished from the background upon excitation.
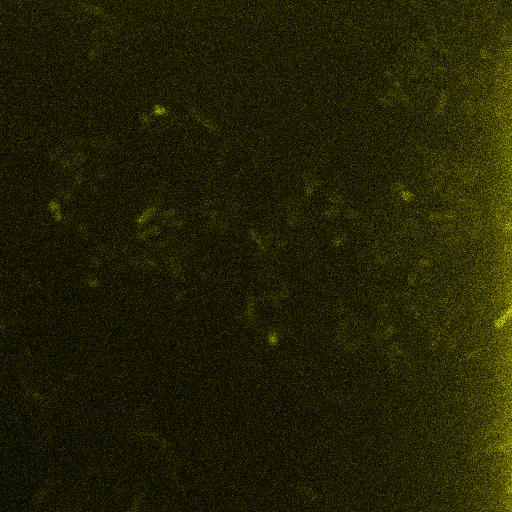
References
[1] Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K. & Miyawaki A. (2002), A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotechnology 20: 87-90.
[2] Marco Weinberg, PhD. Antiviral Gene Therapy Research Unit, Department of Molecular Medicine and Haematology, University of the Witwatersrand Medical School, 7 York Rd. Parktown 2193 SOUTH AFRICA, marc.weinberg@wits.ac.za.
 "
"