Team:Heidelberg/Notebook/miRNA Kit/September
From 2010.igem.org
Laura Nadine (Talk | contribs) |
Laura Nadine (Talk | contribs) |
||
| Line 37: | Line 37: | ||
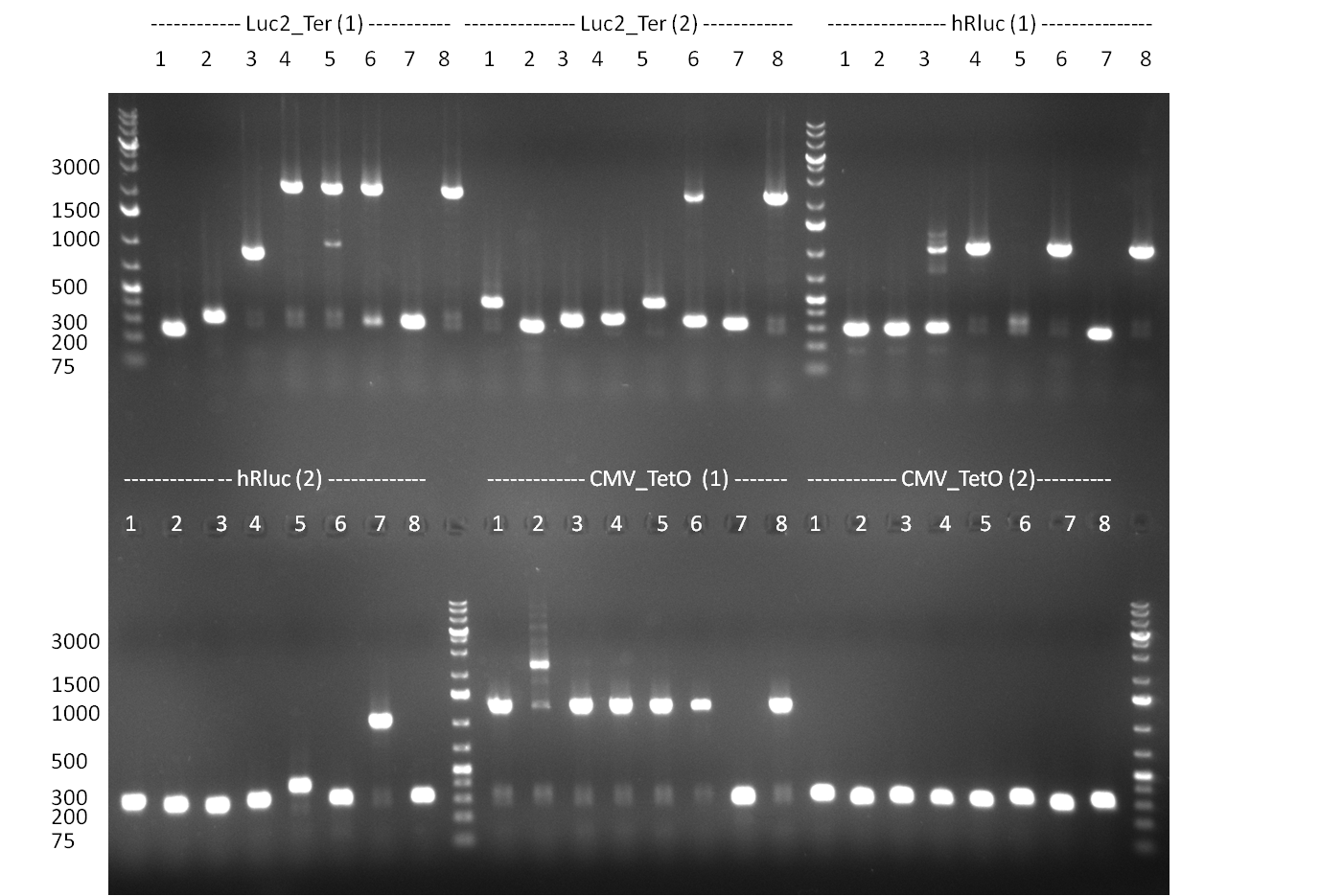
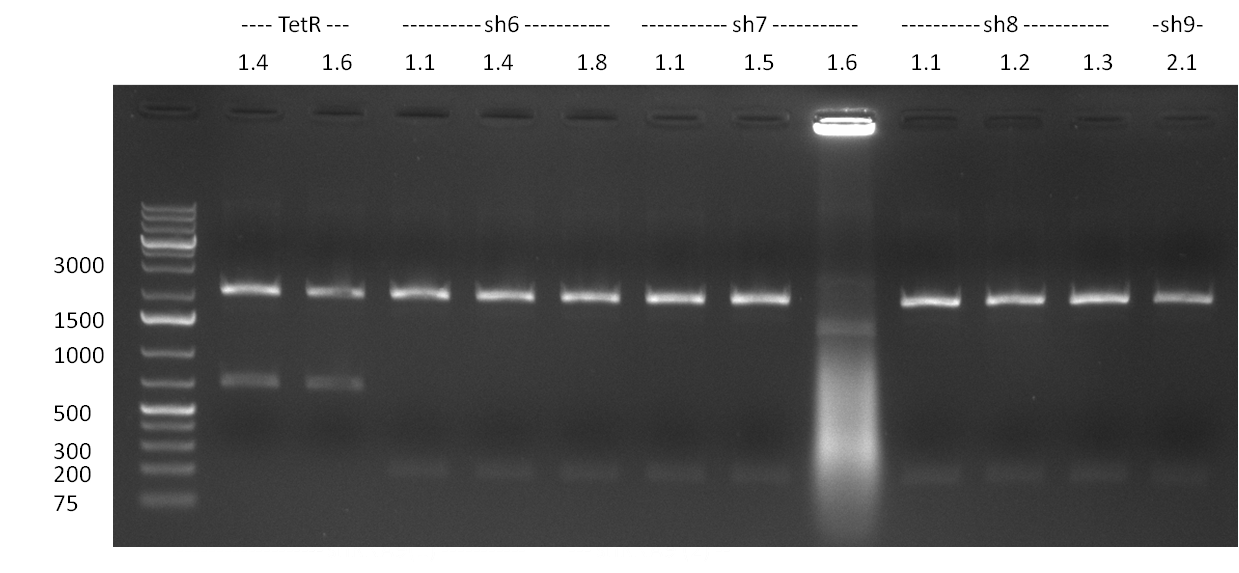
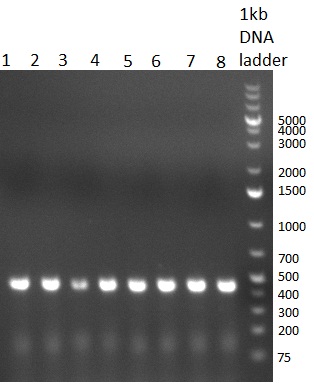
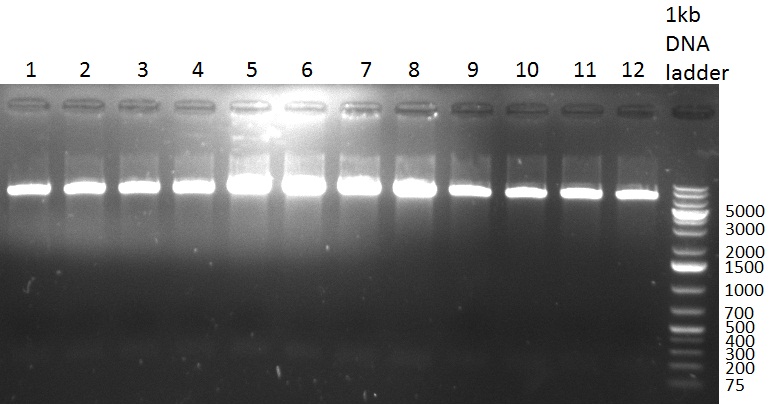
* Test digestion with NotI of the cloned constructs on the previous day (gel 100903-1). For the TetR constructs the expected 600 bp band and for the shRNA constructs, the expected 200 bp band were clearly visible. For each construct, one sample was send for sequencing. Sequencing results were correct for all samples send for sequencing. | * Test digestion with NotI of the cloned constructs on the previous day (gel 100903-1). For the TetR constructs the expected 600 bp band and for the shRNA constructs, the expected 200 bp band were clearly visible. For each construct, one sample was send for sequencing. Sequencing results were correct for all samples send for sequencing. | ||
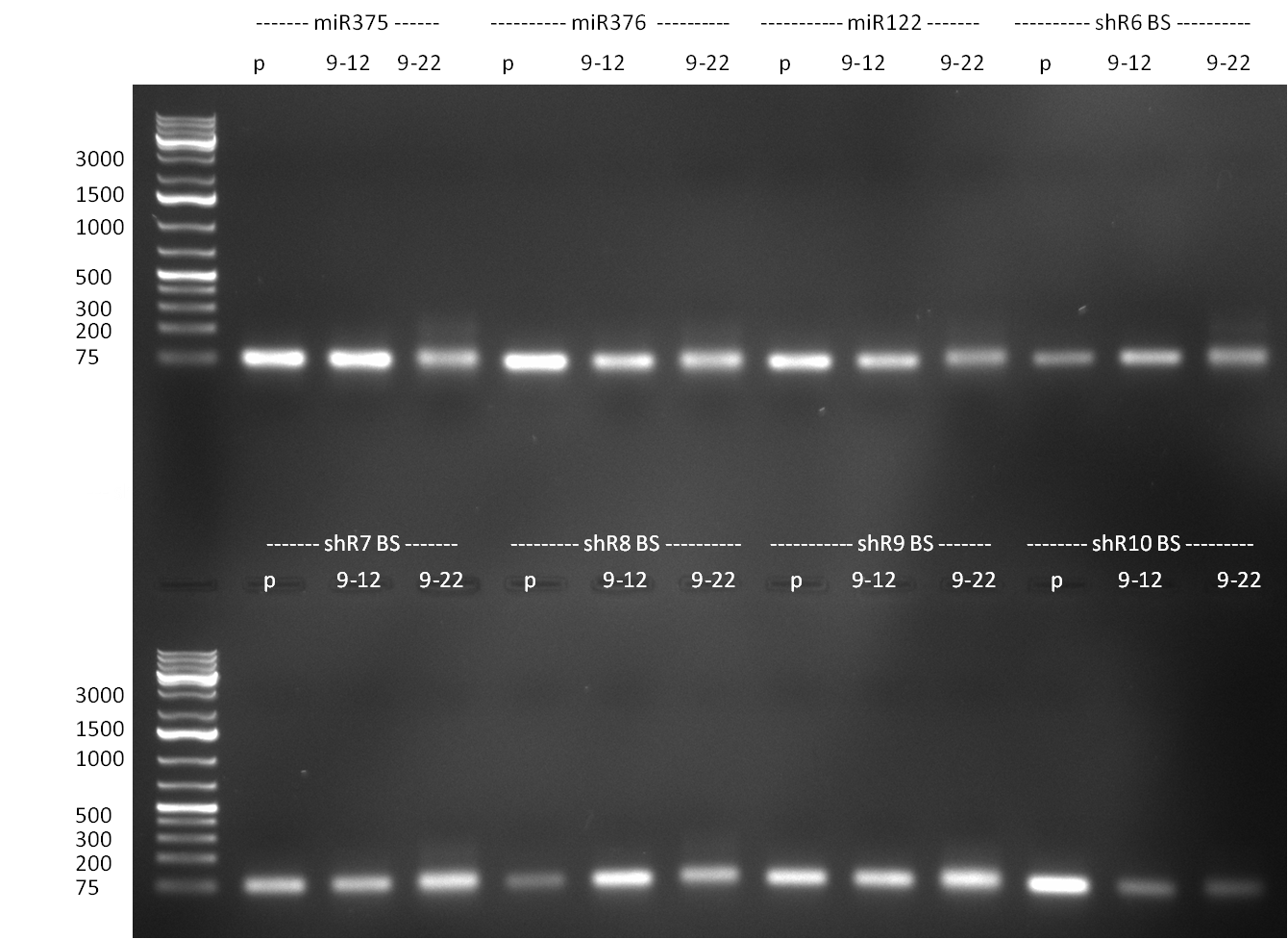
* Purification of single binding sites against shRNA6-10, miR-375/376/122. For each target, the perfect, 9-12 and 9-22 randomized binding sites were purified and analyzed on an agarose gel (100903-2). All bands were clean, with just a slight amount of side product for the 9-22 randomized binding sites. | * Purification of single binding sites against shRNA6-10, miR-375/376/122. For each target, the perfect, 9-12 and 9-22 randomized binding sites were purified and analyzed on an agarose gel (100903-2). All bands were clean, with just a slight amount of side product for the 9-22 randomized binding sites. | ||
| - | <br /> | + | <br /> |
| + | |||
| + | * cloning of shRNAs into pcDNA5/FRT/TO | ||
| + | |||
| + | = 04/09/2010 = | ||
| + | |||
| + | * mini-prep of pcDNA5 with shRNAs 1, 2, 3, 5, 5<nowiki>*</nowiki> and 6 (each four colonies from two independent transformations) | ||
| + | * PCR of shRNA constructs (including shRNA 7, 8 and 9 as well) | ||
| + | ** gel: fine, everywhere amplified sequences below 200 bp | ||
| + | * digestion of promising tuning constructs with BspEI and StuI | ||
| + | ** gel: plasmids seem to be cut only once again (problem maybe BspEI), size is correct | ||
| + | *** suggestion: another digestion on monday with SalI and KpnI, then: submission for sequencing | ||
| + | |||
| + | * mini-prep of pcDNA5 with shRNAs 1, 2, 3, 5, 5<nowiki>*</nowiki> and 6 (each four colonies from two independent transformations) | ||
| + | * PCR of shRNA constructs (including shRNA 7, 8 and 9 as well) | ||
| + | ** gel: fine, everywhere amplified sequences below 200 bp | ||
| + | * digestion of promising tuning constructs with BspEI and StuI | ||
| + | ** gel: plasmids seem to be cut only once again (problem maybe BspEI), size is correct | ||
| + | *** suggestion: another digestion on monday with SalI and KpnI, then: submission for sequencing | ||
| + | |||
= 05/09/2010 = | = 05/09/2010 = | ||
| Line 48: | Line 67: | ||
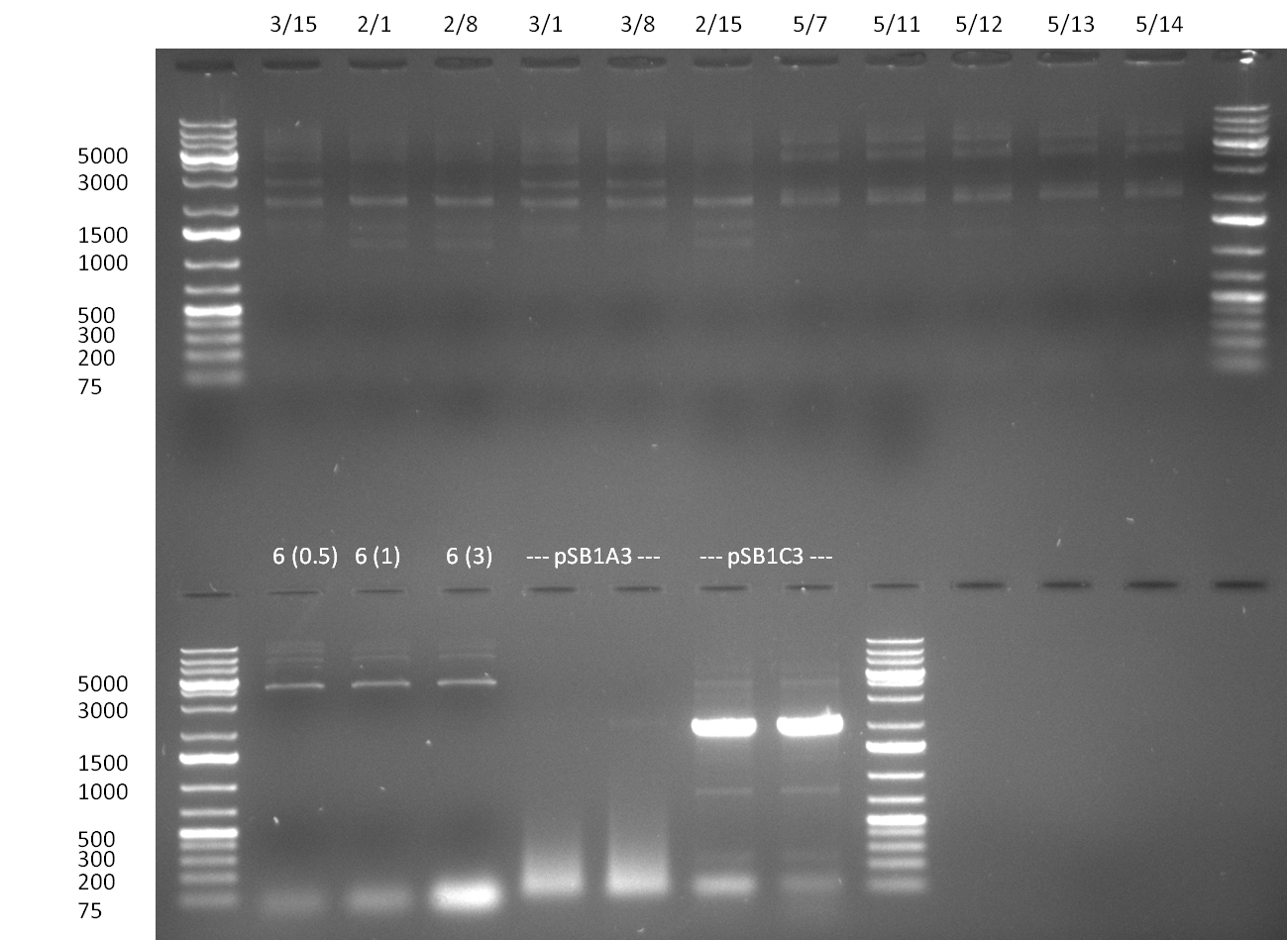
* digestion of PCR amplified vector backbone pSB1A3 with EcoRI/PstI; after subsequent heat inactivation, digestion with DpnI was performed and the vector was purified by applying a nucleotide removal kit. | * digestion of PCR amplified vector backbone pSB1A3 with EcoRI/PstI; after subsequent heat inactivation, digestion with DpnI was performed and the vector was purified by applying a nucleotide removal kit. | ||
<br /> | <br /> | ||
| + | * digestion of tuning construct with KpnI and SalI result in weird band pattern on the gel | ||
| + | * PCR and purification of the binding sites looked fine on the gel | ||
| + | * submission of tuning constructs and pcDNA5 containing different miRNAs for sequencing | ||
| + | |||
<br /> | <br /> | ||
= 07/09/2010 = | = 07/09/2010 = | ||
| Line 66: | Line 89: | ||
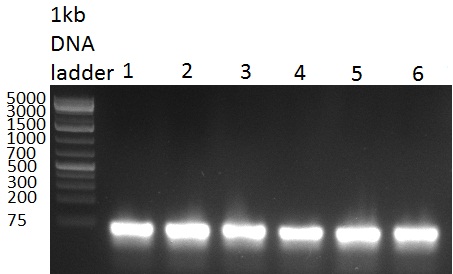
All Ligations were controlled by loading 10 ul of ligation reactiong onto a 1 % agarose gel (100907-1), run for 35 min @ 100 V | All Ligations were controlled by loading 10 ul of ligation reactiong onto a 1 % agarose gel (100907-1), run for 35 min @ 100 V | ||
<br /> | <br /> | ||
| + | * repetition of TetR and tuning construct cloning | ||
| + | * purification and digestion of PCR amplified binding sites with NotI | ||
| + | |||
<br /> | <br /> | ||
| Line 72: | Line 98: | ||
<br /> | <br /> | ||
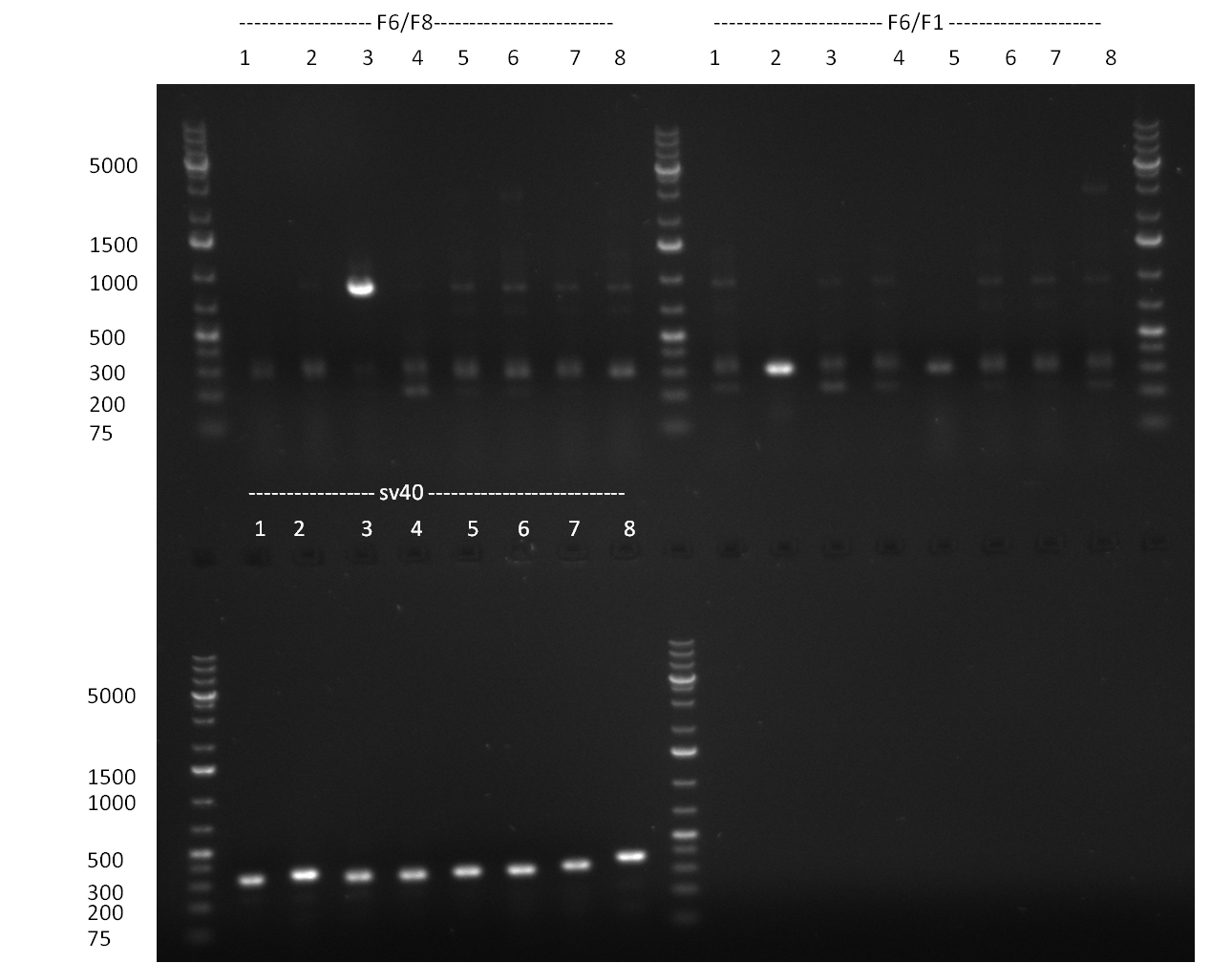
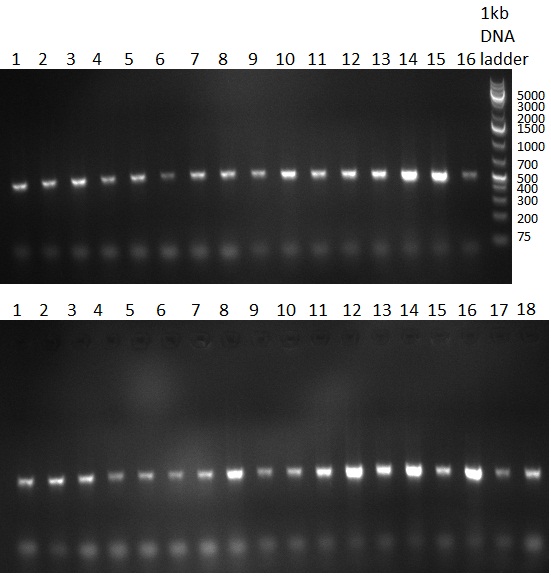
* Colony PCR of the previous days' cloning and miniprep cultures were inocculated in parallel; The result of the PCR was analyzed on a 1 % agarsoe gel (100908-1) run for 35 min @ 100 V. | * Colony PCR of the previous days' cloning and miniprep cultures were inocculated in parallel; The result of the PCR was analyzed on a 1 % agarsoe gel (100908-1) run for 35 min @ 100 V. | ||
| + | |||
| + | * cloning of shRNAs in pcDNA5 again | ||
| + | * further digestion of binding sites with AscSI (analog to SgfI) and purification | ||
| + | * preparation of psyCHECK2 for insertion of binding sites and shRNAs for first test measurements | ||
| + | * transformation of TetR constructs | ||
| + | * ligation of both inserts for the tuning construct, extraction of the right insert (3000bp) and ligation into the backbone | ||
| + | |||
<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
| Line 80: | Line 113: | ||
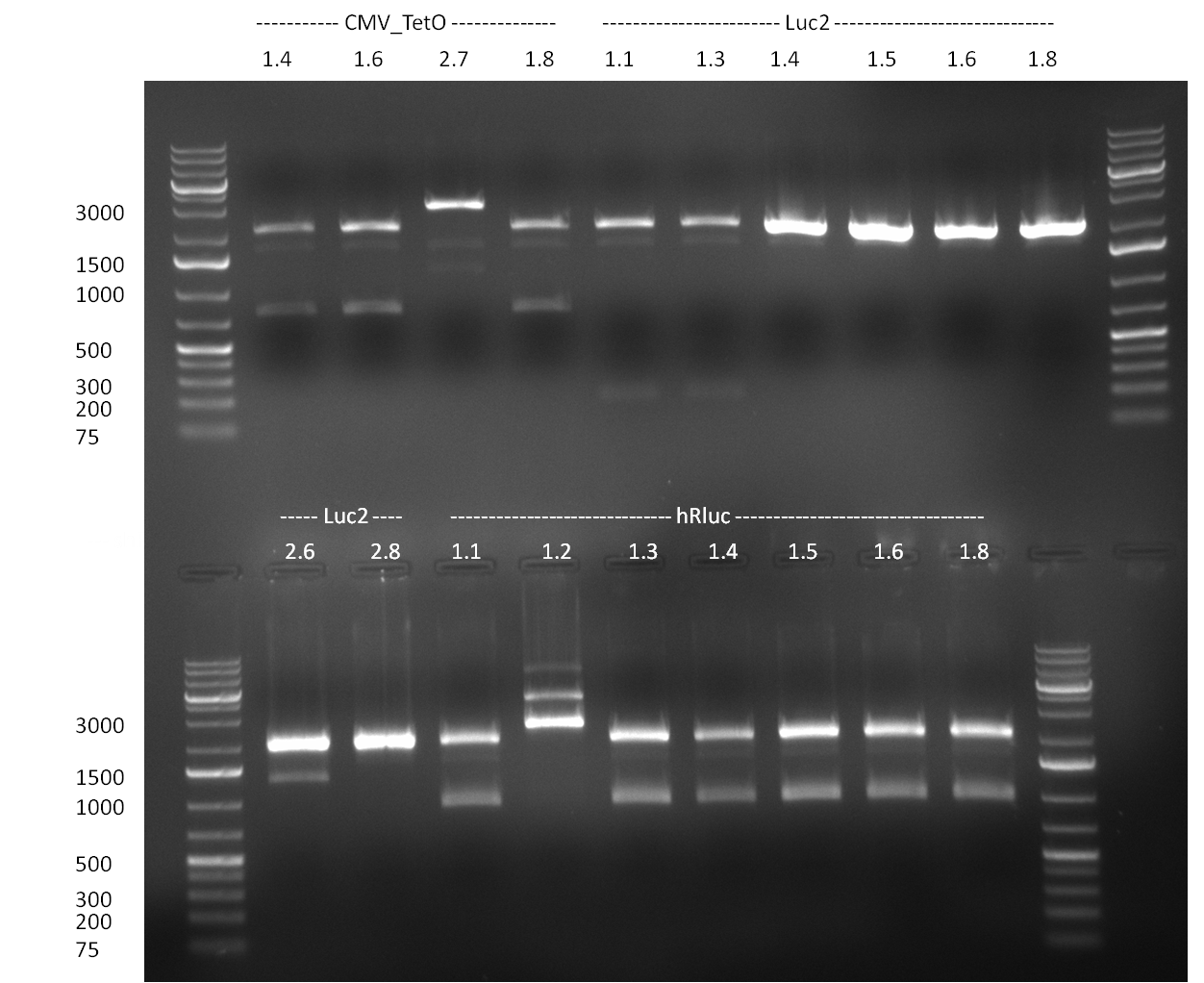
*The nine Luc2-SV40 constructs were with XbaI and NheI (negative testing), and XhoI and PstI (positive testing) for the induced mutation. The reaction was set up according to the standard protocol, and then the samples were visualized on a 1% agarose gel (gel 100909-2). | *The nine Luc2-SV40 constructs were with XbaI and NheI (negative testing), and XhoI and PstI (positive testing) for the induced mutation. The reaction was set up according to the standard protocol, and then the samples were visualized on a 1% agarose gel (gel 100909-2). | ||
| + | * colony PCR of TetR constructs are positive | ||
| + | * inoculate TetR for miniprep | ||
<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
<br /><br /> | <br /><br /> | ||
| Line 97: | Line 132: | ||
Subsequent Transformation into DH5alpha cells | Subsequent Transformation into DH5alpha cells | ||
<br /> | <br /> | ||
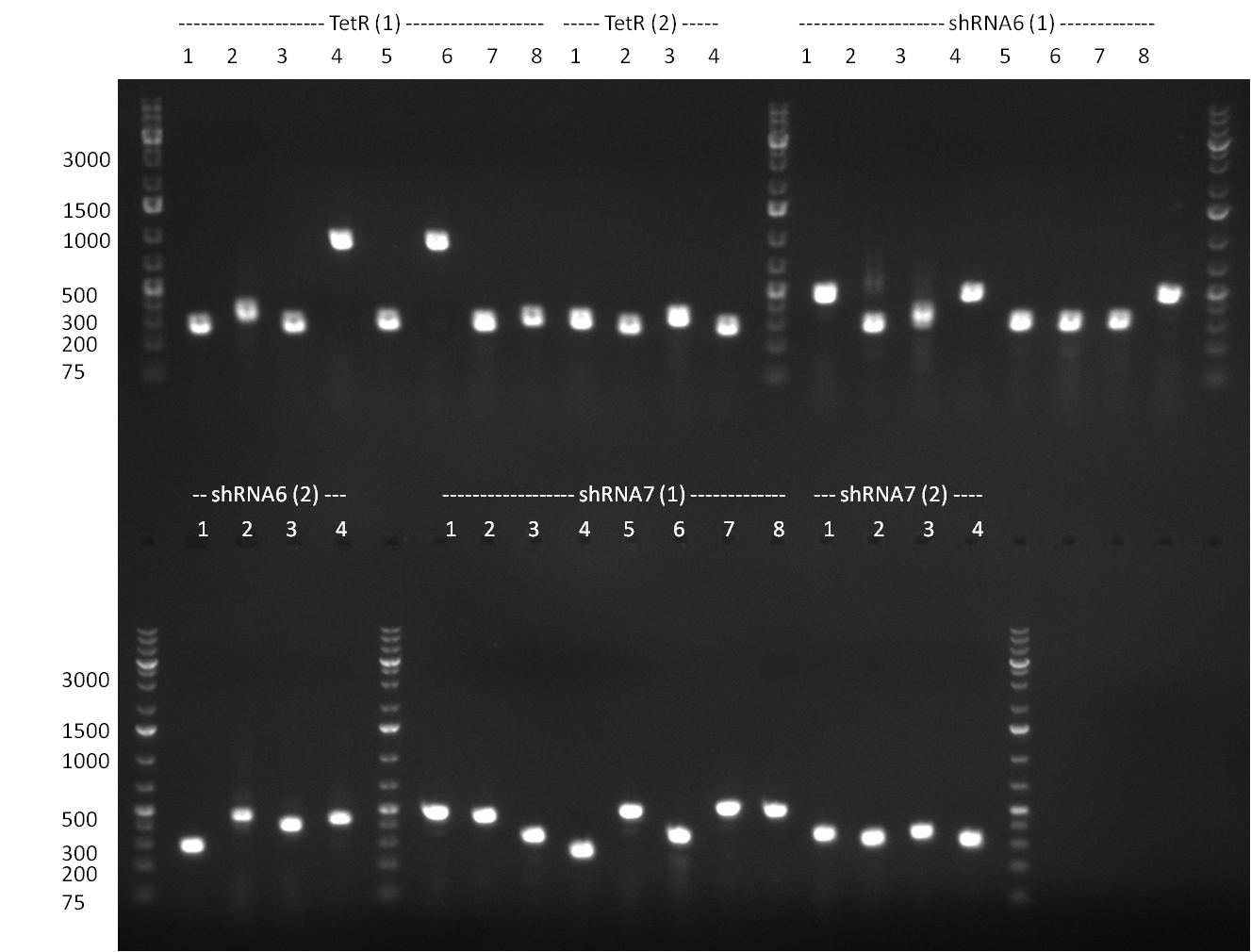
| + | * miniprep of TetR | ||
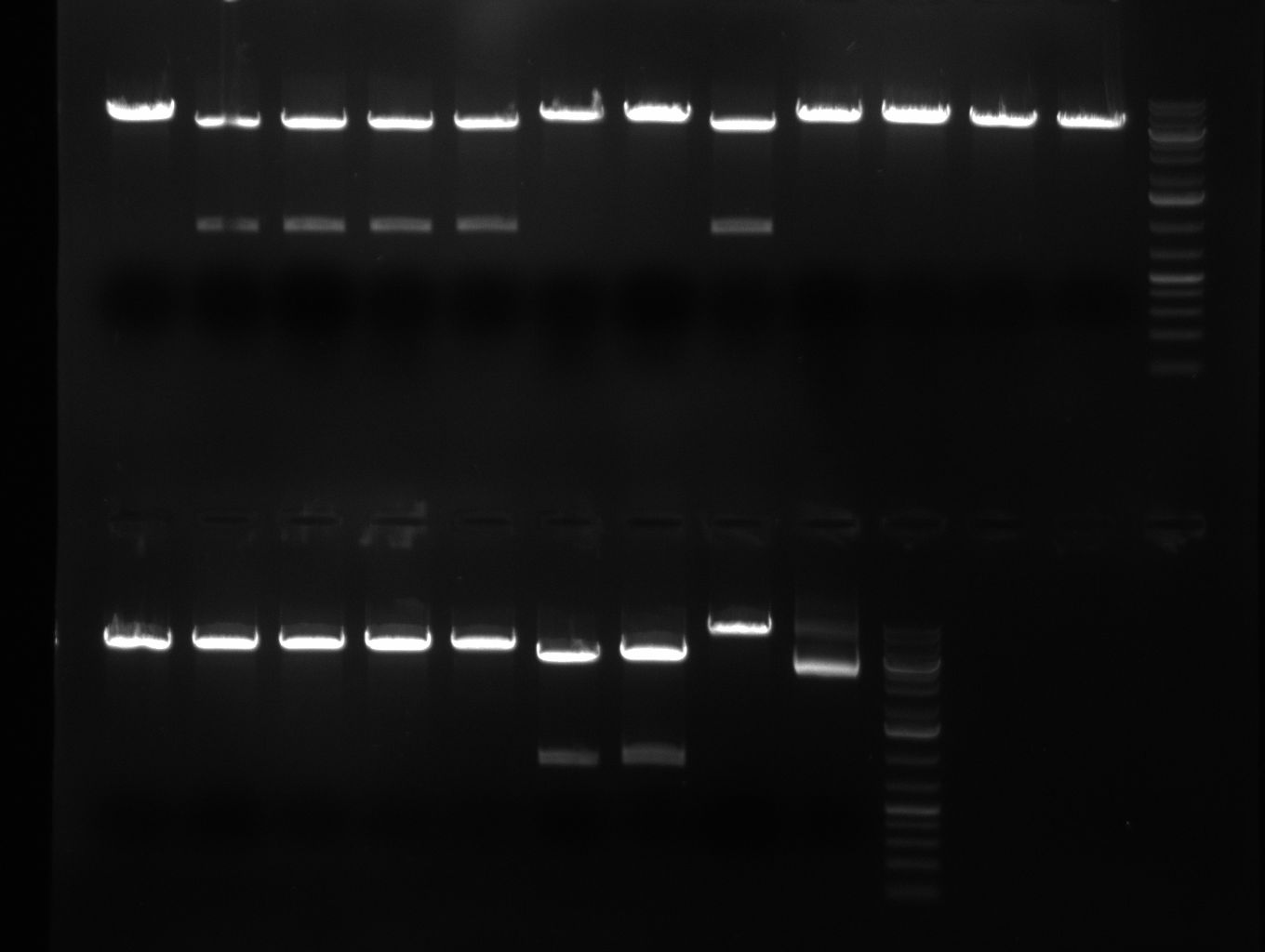
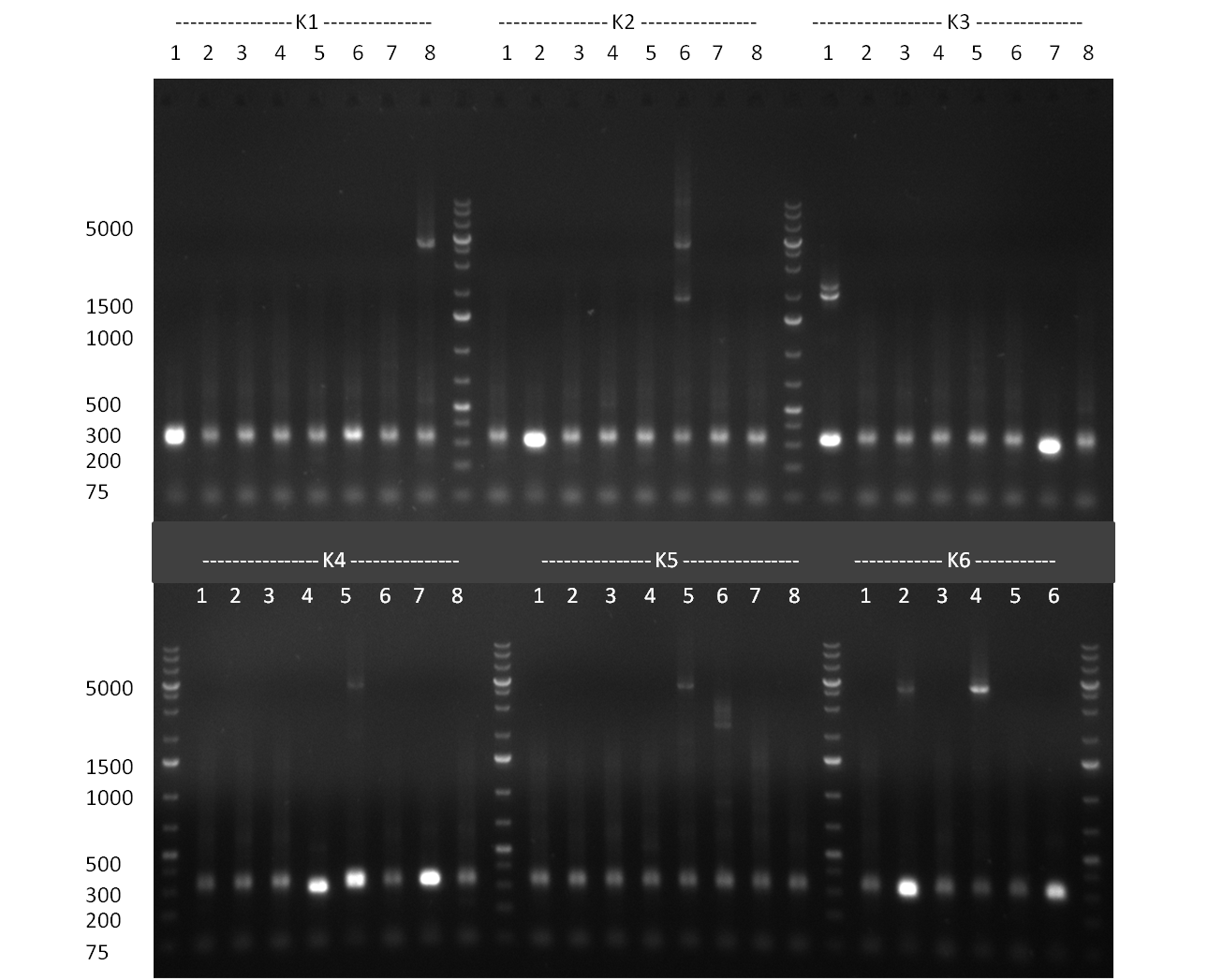
| + | * test digest of TetR with BspEI & NotI and with SgfI gave promising results | ||
| + | ** YES it was done with marker instead of loading dye but: | ||
| + | *** for lanes 2-4 you see a stronger band at 700 pb (size of TetR) | ||
| + | *** for landes 5-7 you see a stronger band at 5000 bp as it was linearized with SgfI | ||
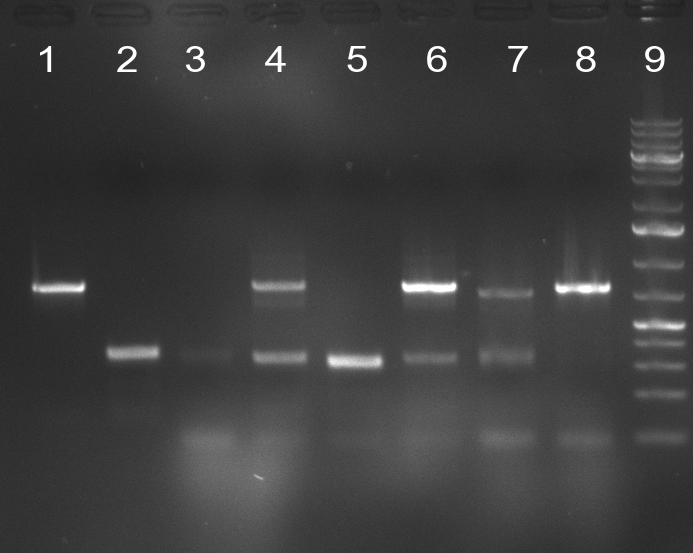
| + | [[Image:100910-rbtettestdigest.jpg|350px]] | ||
| + | * colony PCR of binding site in PsiCheck and tuning construct.. tuning construct looks promising, BS nothing on the gel except for primers | ||
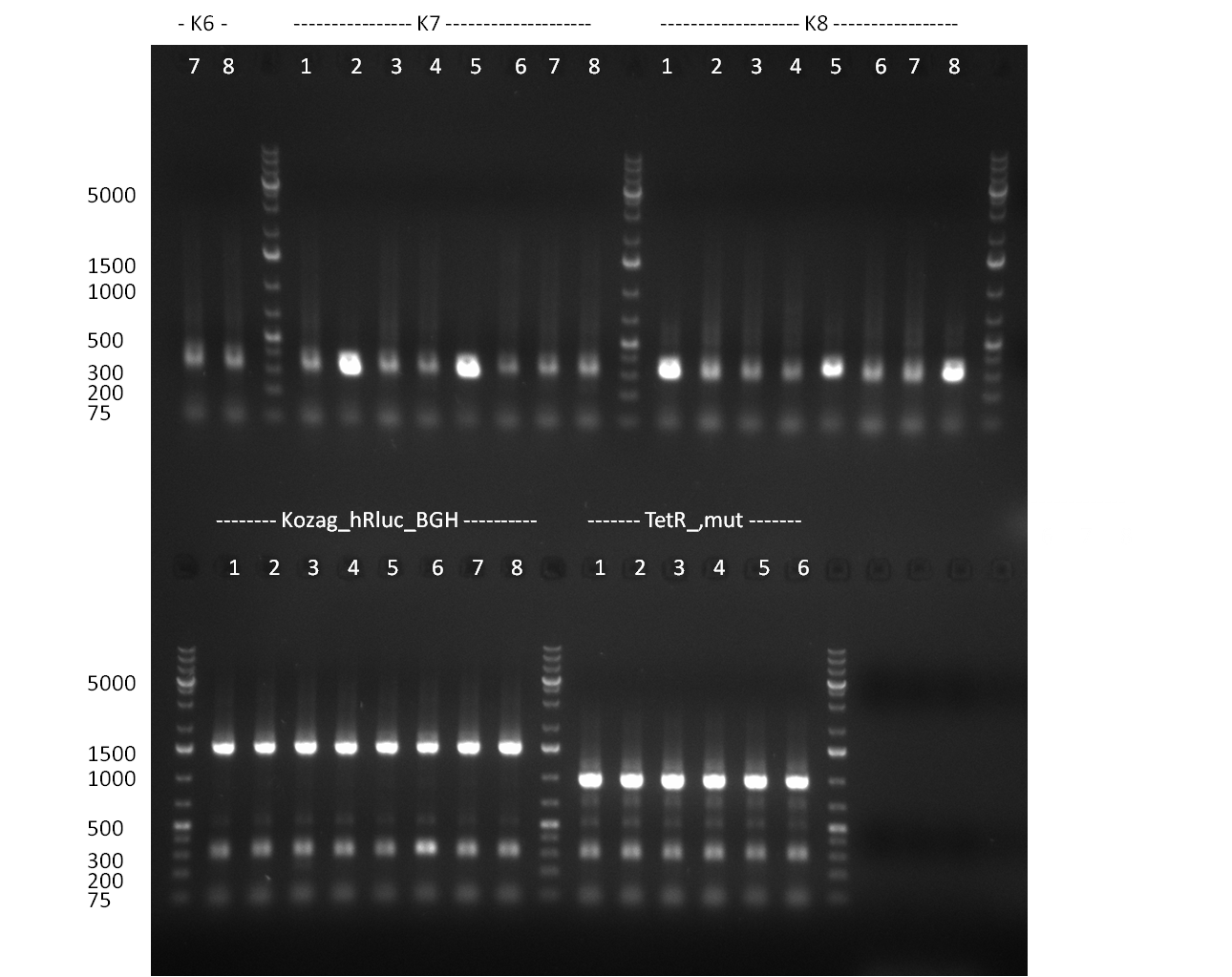
| + | * colony PCR of shRNAs look promising even though we cannot say whether still the old shRNA is inside | ||
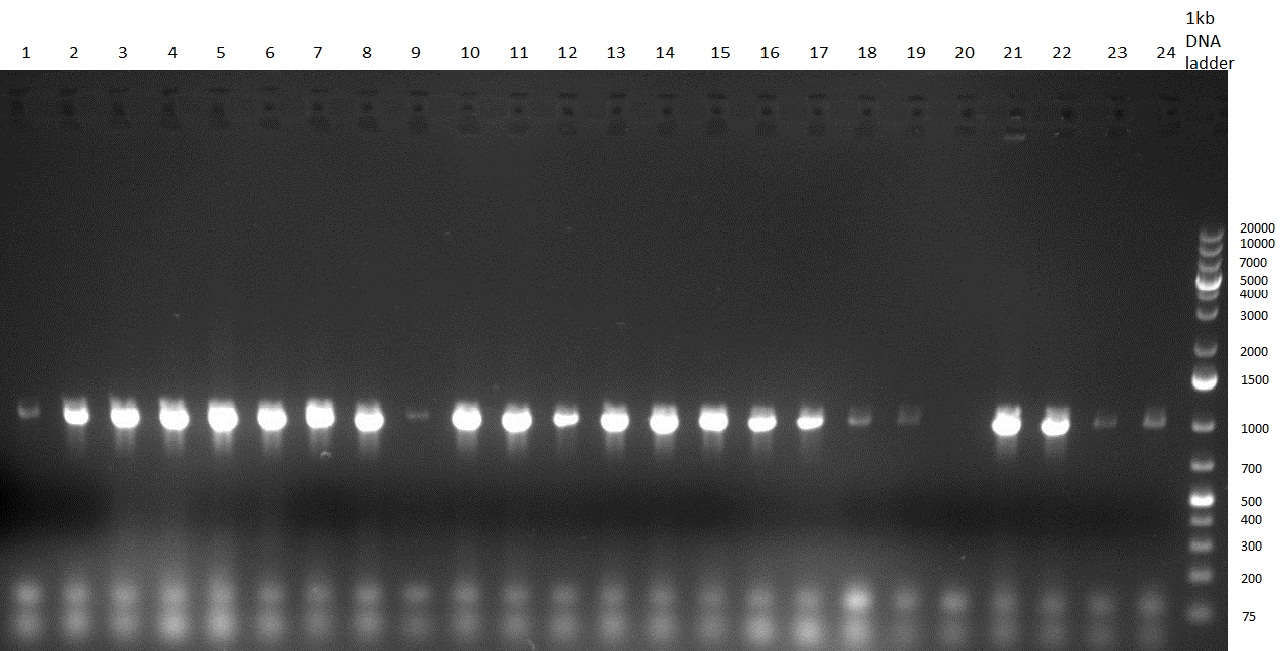
| + | ** PCR of CMV shRNA which leads to a size of 1200bp | ||
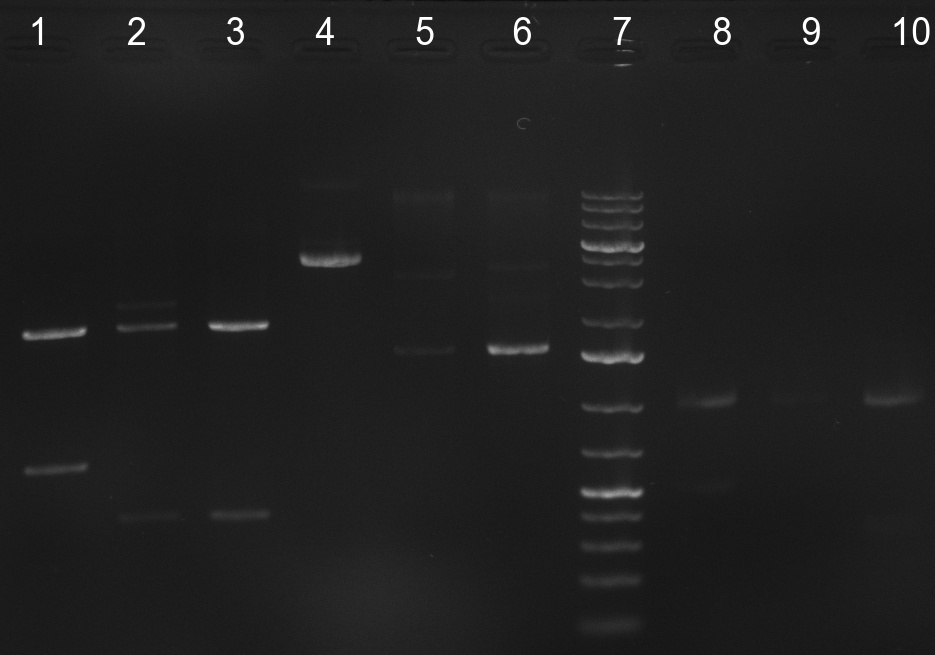
| + | [[Image:Rb100910colonyshRNAs.jpg|350px]] | ||
| + | |||
| + | |||
= 11/09/2010 = | = 11/09/2010 = | ||
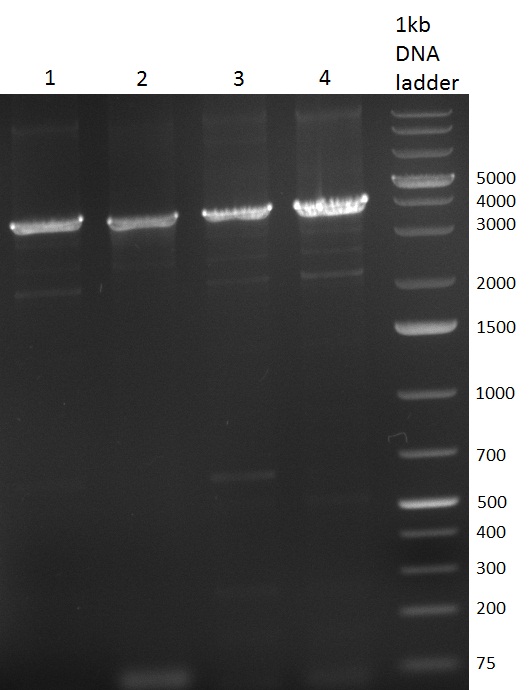
[[Image:20100911_gel1.png|thumb|350 px|right|Gel 100911-1]] | [[Image:20100911_gel1.png|thumb|350 px|right|Gel 100911-1]] | ||
| Line 102: | Line 149: | ||
<br /> | <br /> | ||
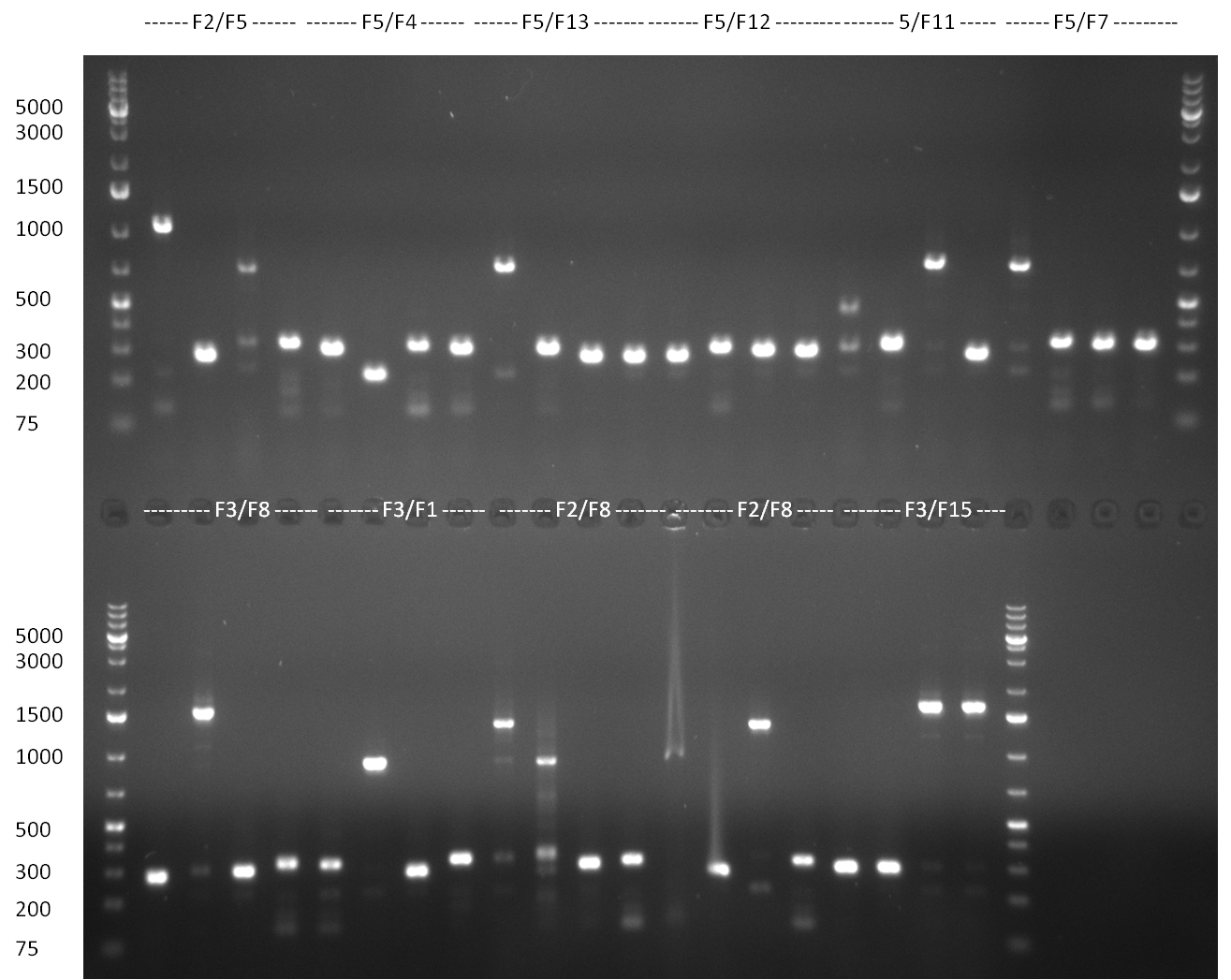
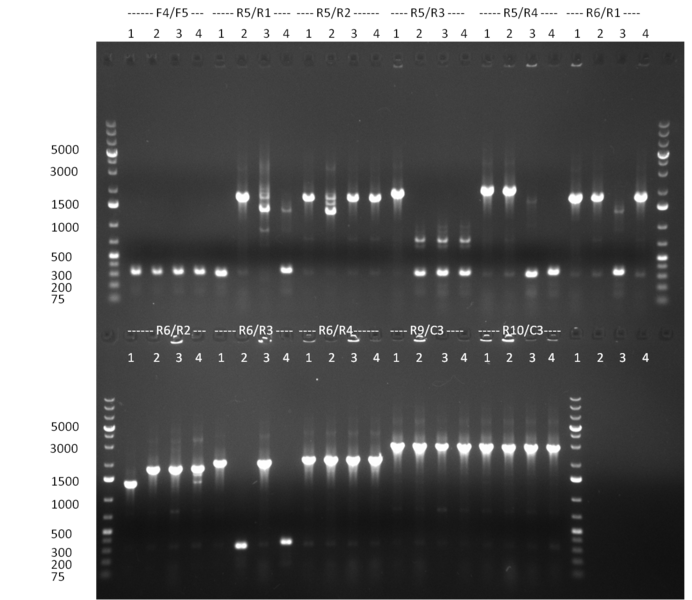
* Colonies were picked from the plates from the previous days' cloning and a colony PCR was performed according to the standard protocol. In parallel LB miniprep cultures were inocculated for each colony (gel 100911-1 and 100911-2). | * Colonies were picked from the plates from the previous days' cloning and a colony PCR was performed according to the standard protocol. In parallel LB miniprep cultures were inocculated for each colony (gel 100911-1 and 100911-2). | ||
| - | + | ||
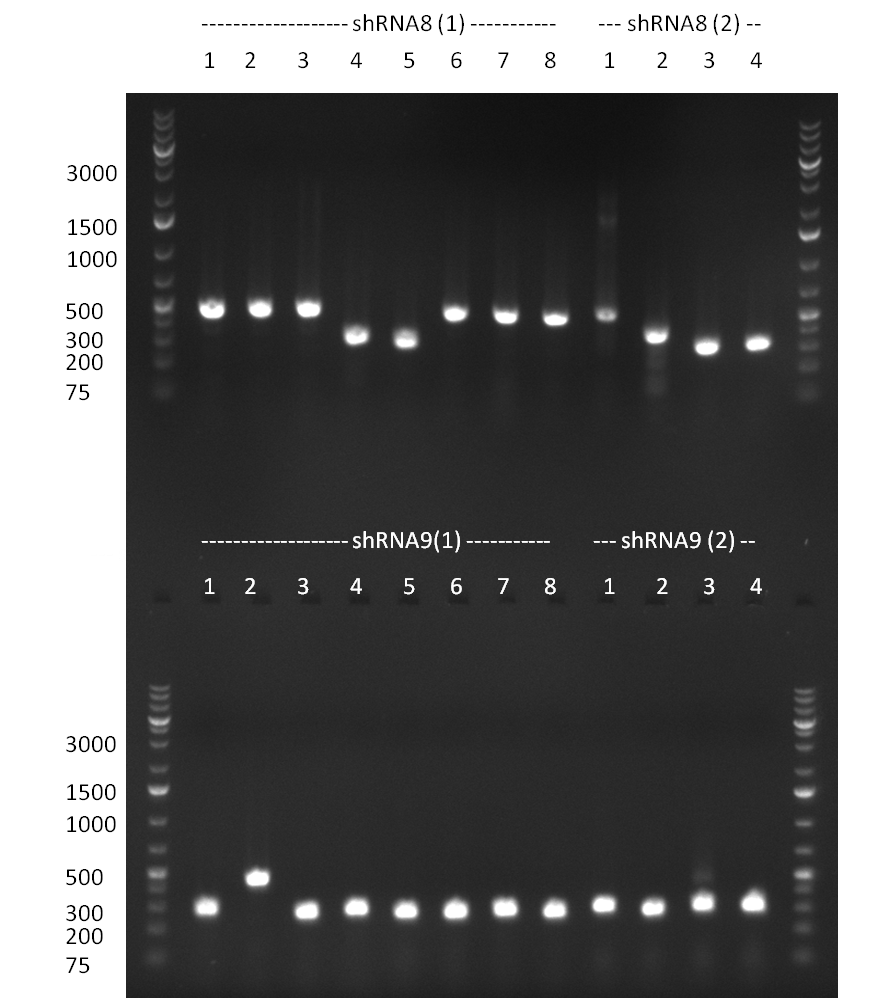
| - | + | * miniprep of tuning construct and assorted shRNA BS PsiCheck colonies. Repetition of test PCR (for tuning primer L11 and L13, for BS primer L1 and 86a (dominik)) with miniprep template. Tuning construct looks kind of good on the gel, but I don't want to get you hopes up too soon.. it will be send for sequencing on monday. | |
| - | + | * Tet Repressor cloned!!! | |
| - | + | * shRNA 2,3,5,7,8 and 9 seems to be cloned according to test digest | |
| - | + | [[Image:Rb100911_testdigestshRNA.jpg|350px]] | |
| + | |||
= 12/09/2010 = | = 12/09/2010 = | ||
| Line 113: | Line 161: | ||
<br /> | <br /> | ||
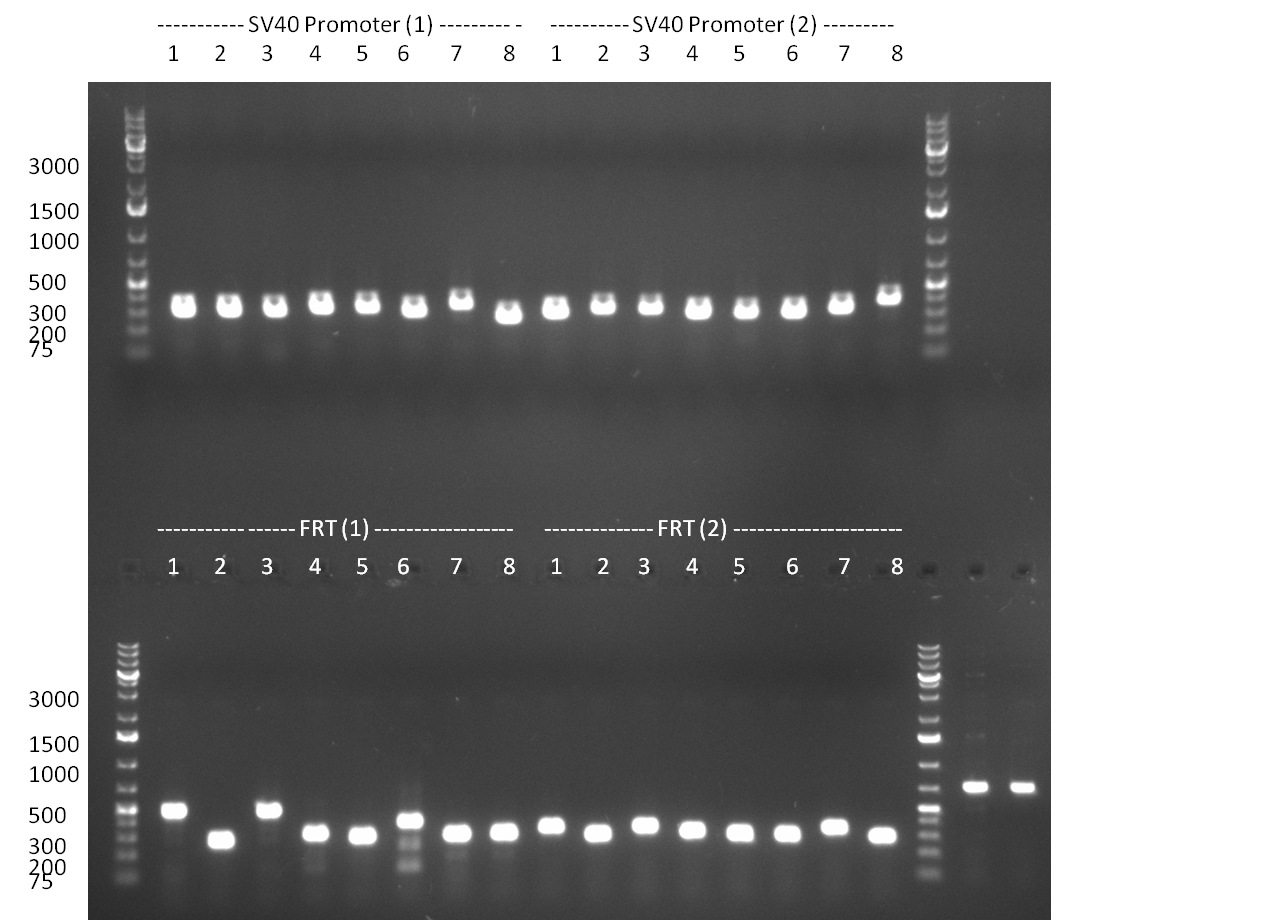
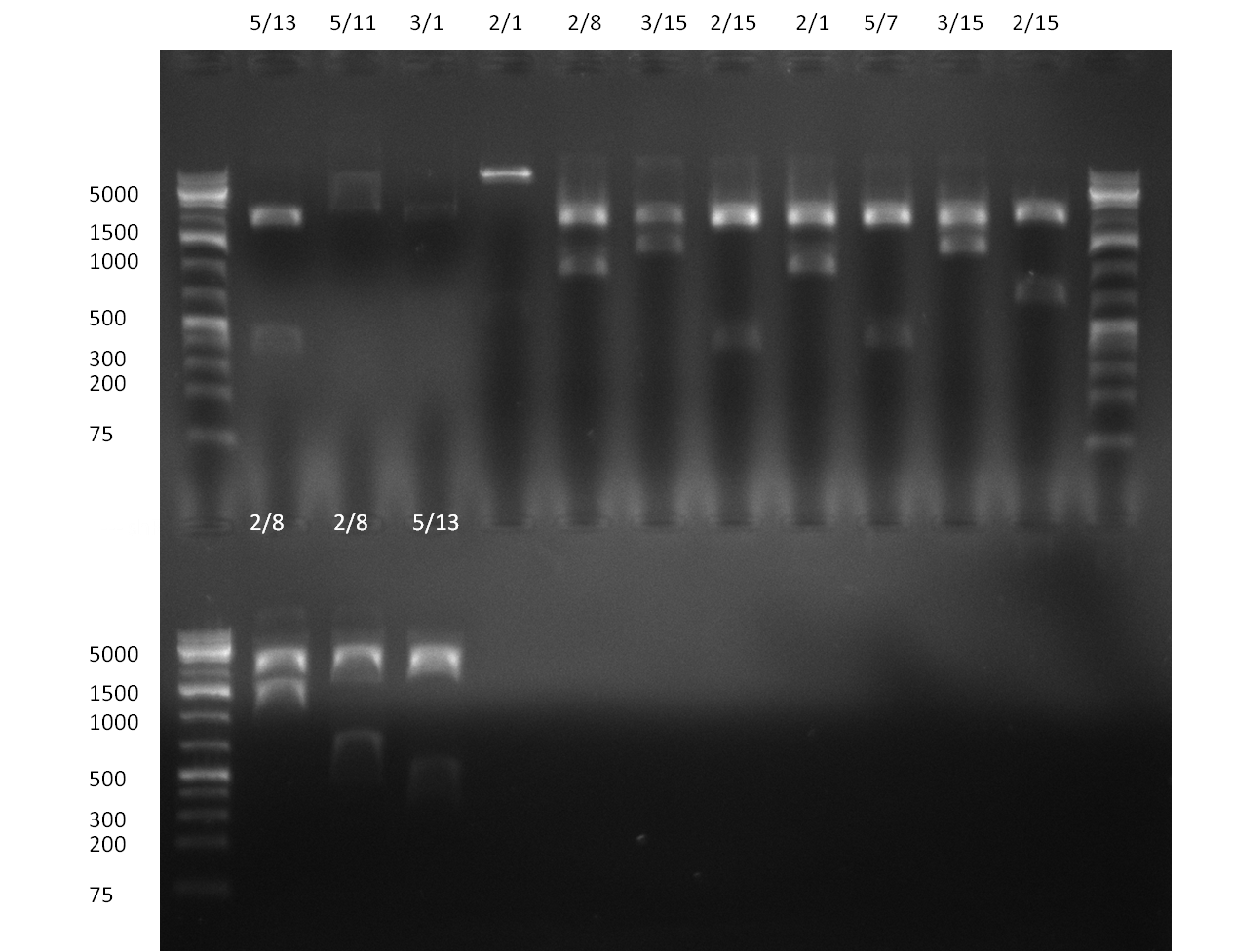
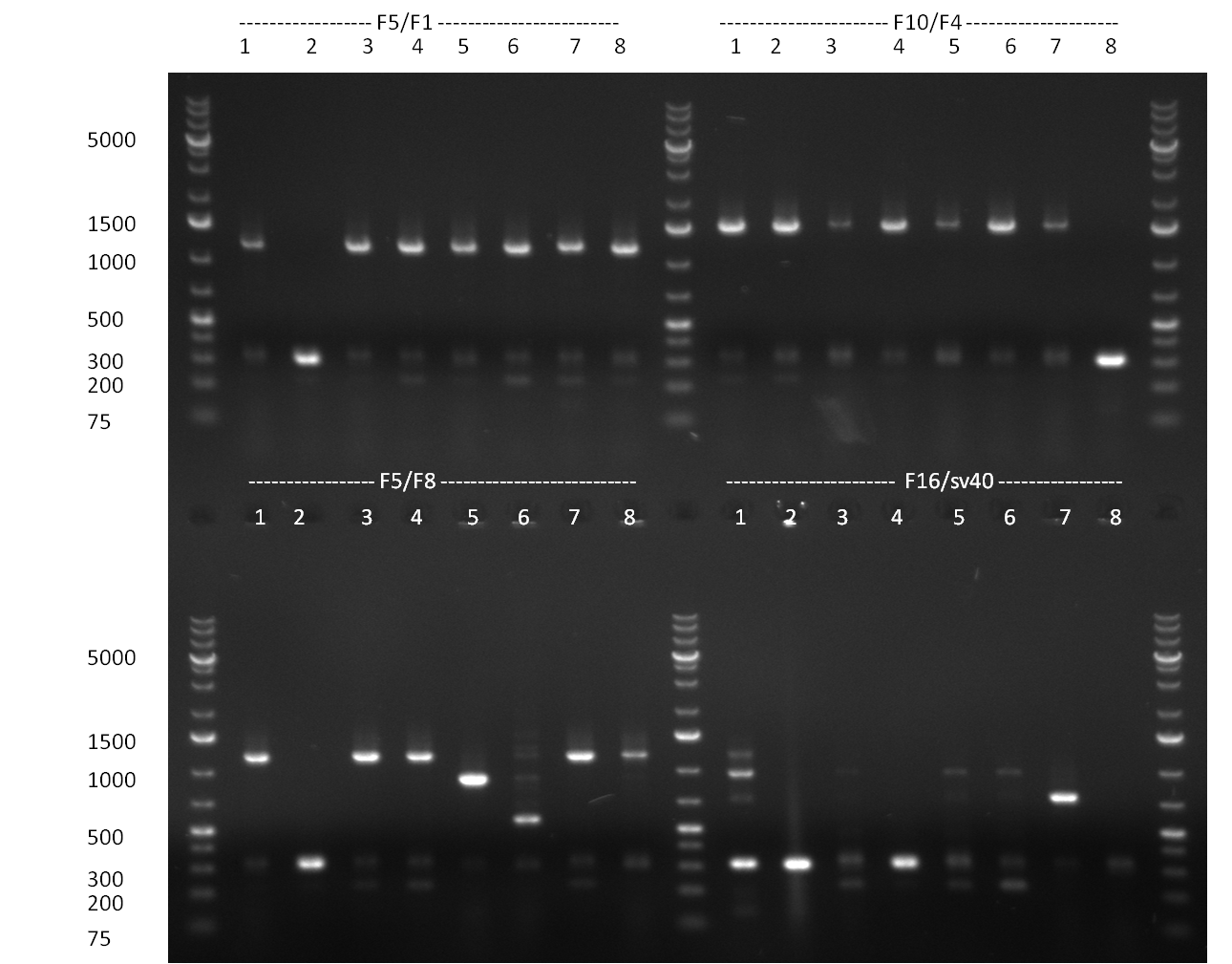
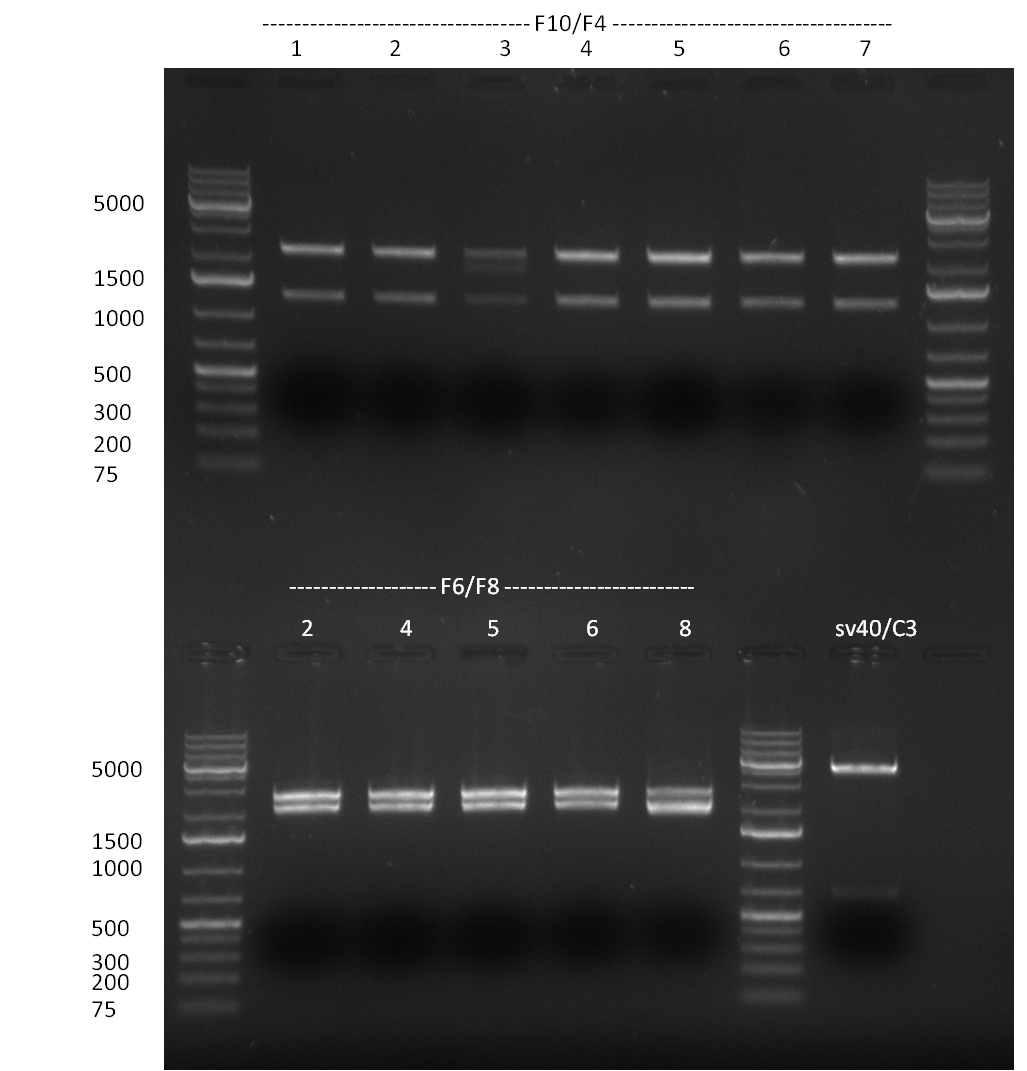
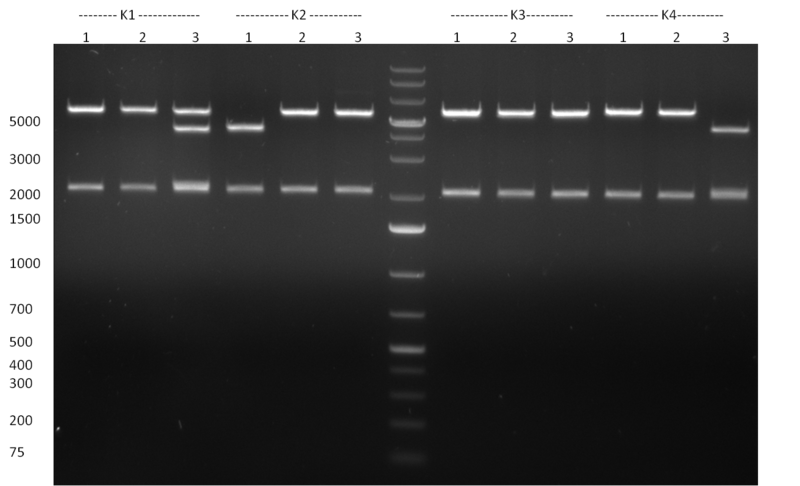
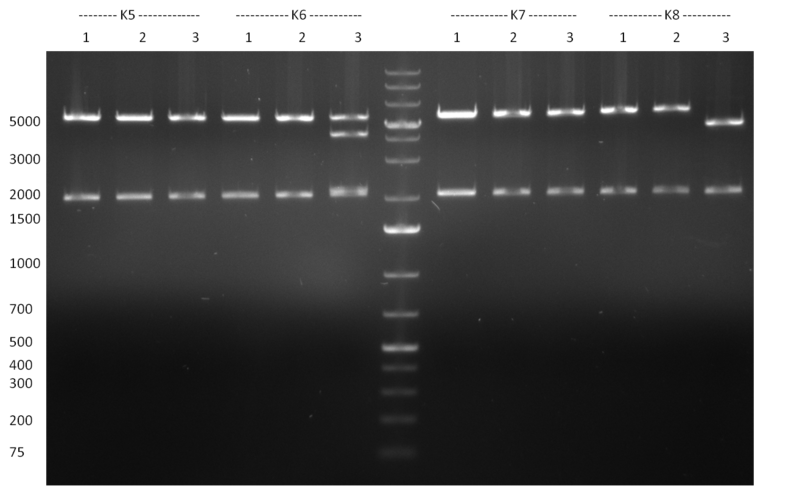
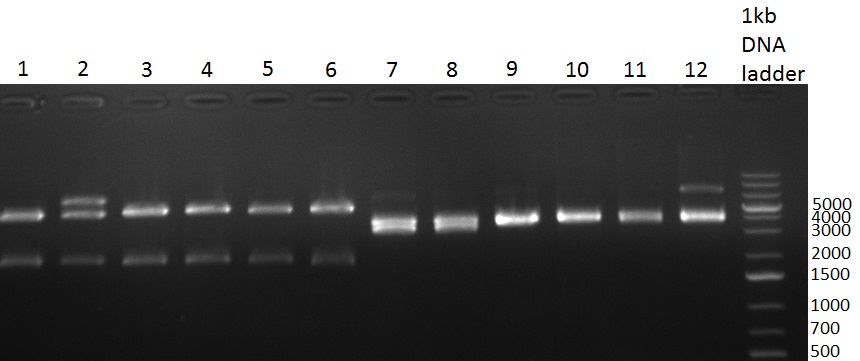
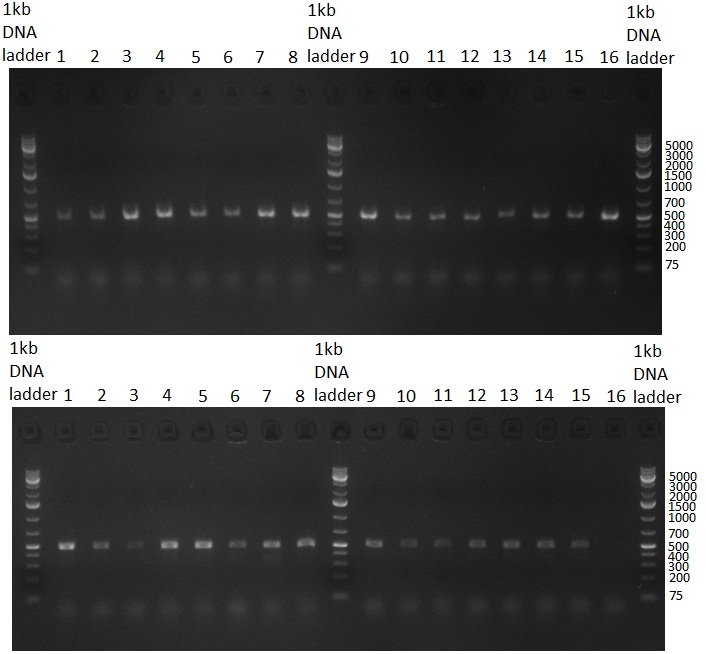
* Minipreps were done by applying a Qiagen Miniprep Kit for the positive candidates picked on the previous day. Test digestion was performed by digesting ~ 250 ng of each construct with SpeI/NheI and the result was analyzed on a 1 % agarose gel (100912-1 and100912-2) run for 40 min @ 100 V. For all the constructs we got the right band despite construct F10/F4.3. Positive constructs were send for sequencing @ GATC. | * Minipreps were done by applying a Qiagen Miniprep Kit for the positive candidates picked on the previous day. Test digestion was performed by digesting ~ 250 ng of each construct with SpeI/NheI and the result was analyzed on a 1 % agarose gel (100912-1 and100912-2) run for 40 min @ 100 V. For all the constructs we got the right band despite construct F10/F4.3. Positive constructs were send for sequencing @ GATC. | ||
| + | |||
| + | * miniprep of shRNA constructs and Tuning constructs | ||
<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
| Line 129: | Line 179: | ||
* transformation of 10 ul ligation reaction after heat inactivation into DH5alpha cells | * transformation of 10 ul ligation reaction after heat inactivation into DH5alpha cells | ||
| - | + | * digestion and cloning of binding sites and PsiCheck.. again | |
| + | * submission of constructs for sequencing | ||
| + | |||
| + | |||
= 14/09/2010 = | = 14/09/2010 = | ||
[[Image:20100914_HD10.png|thumb|350 px|right|Gel 100914-1]] | [[Image:20100914_HD10.png|thumb|350 px|right|Gel 100914-1]] | ||
[[Image:20100914_gel2_HD10.png|thumb|350 px|right|Gel 100914-2]] | [[Image:20100914_gel2_HD10.png|thumb|350 px|right|Gel 100914-2]] | ||
<br /> | <br /> | ||
| + | * '''note''': for sequential digest of pcDNA5 or shRNAs with ApaI and HindIII use buffer 2 | ||
| + | |||
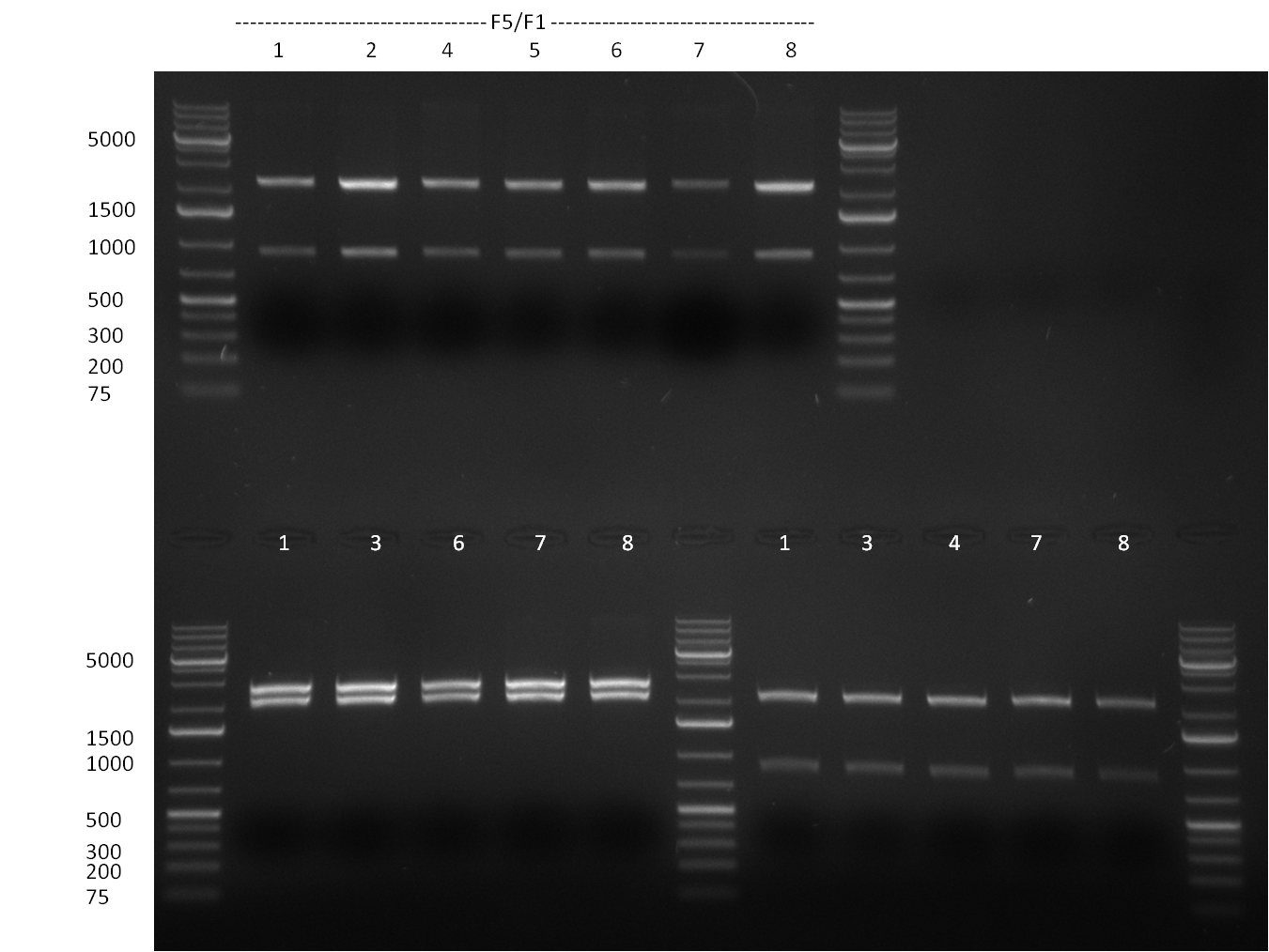
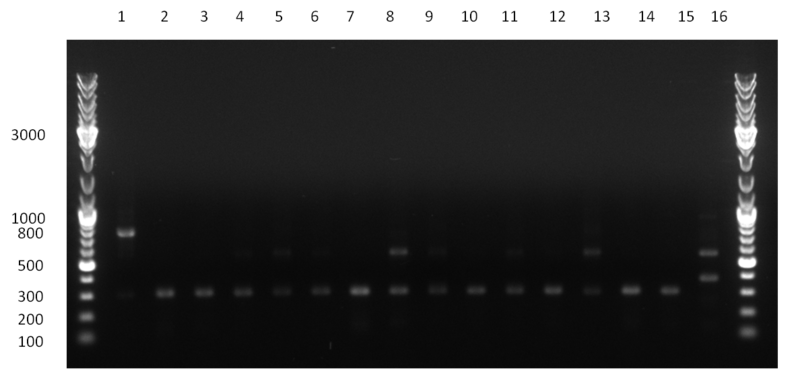
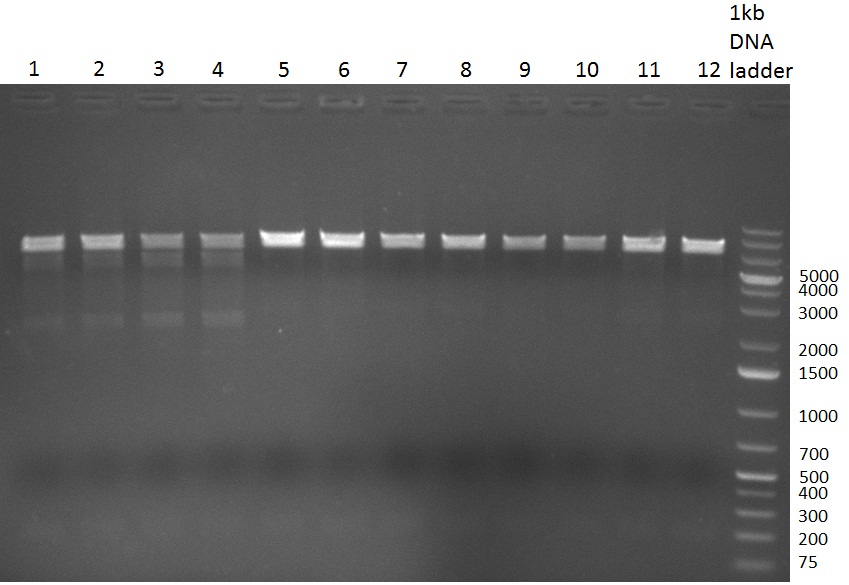
* Colony- PCR for the cloning products done on the previous day (gels 100914-1 and 100914-2 on the right) | * Colony- PCR for the cloning products done on the previous day (gels 100914-1 and 100914-2 on the right) | ||
* positive clones were Mini-Prepped and test digested | * positive clones were Mini-Prepped and test digested | ||
| Line 140: | Line 195: | ||
= 15/09/2010 = | = 15/09/2010 = | ||
[[Image:20100915_gel1_HD2010.png|thumb|350px|right| Gel 100915-1]] | [[Image:20100915_gel1_HD2010.png|thumb|350px|right| Gel 100915-1]] | ||
| + | <br /> | ||
| + | * digestion of TetR construct with AsiSi and NotI or SgfI and NotI respectively | ||
| + | ** for cloning of binding sites for miR122, miR375 and miR376 | ||
| + | * ligation of the other binding sites for synthetic pri-miRNA (6, 7, 8, 9 and 10) with digested psiCHECK2 | ||
<br /> | <br /> | ||
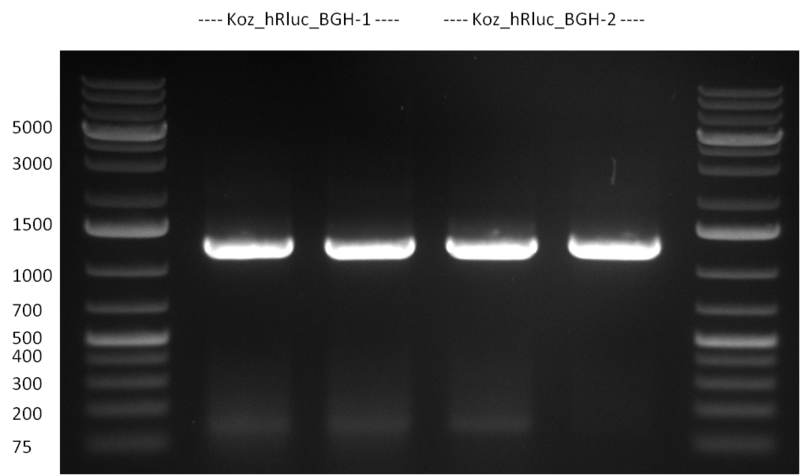
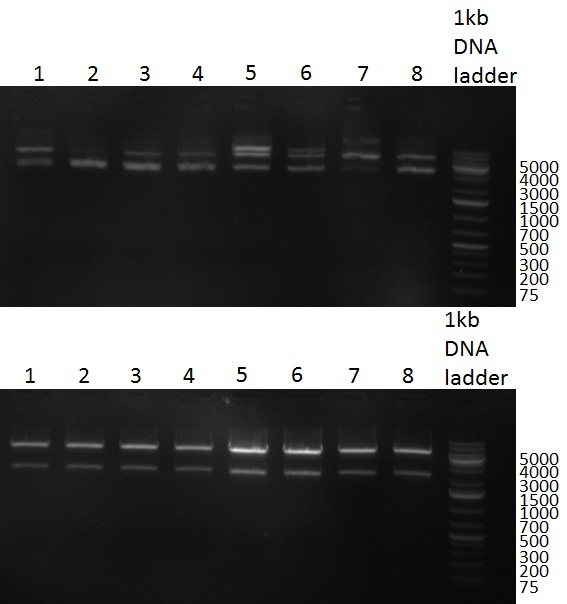
* PCR for the introduction of the Kozag-Sequence in front of the hRluc_BGH part; two strategies were performed in parallel using different touchdown-PCR protocols. Protocol nr. 1 was set up as follows: | * PCR for the introduction of the Kozag-Sequence in front of the hRluc_BGH part; two strategies were performed in parallel using different touchdown-PCR protocols. Protocol nr. 1 was set up as follows: | ||
| Line 196: | Line 255: | ||
:* Ligation and Transformation were done according to the standard protocol | :* Ligation and Transformation were done according to the standard protocol | ||
<br /> | <br /> | ||
| + | |||
| + | * PCR to generate binding site oligos, nucleotid removal, digestion, nucleotid removal | ||
| + | * different digestion and ligation protocols tested for binding sites and psiCHECK2, afterwards transformation | ||
| + | * possibilities: | ||
| + | ** dilution of oligo insert <nowiki>[1:100,1:500]</nowiki>, ratio: insert/vector = 5/1 | ||
| + | ** SAP treatment for oligos to prevent them from forming concatemers | ||
| + | ** SAP treatment of backbone to prevent it from re-ligation | ||
| + | ** cut of backbone into two fragments to ease gel excision (if done) and advance ligation | ||
| + | ** ligation process: 1h @ RT or over-night @ 4°C in darkness | ||
| + | *** note: bs122 confused with bs7 | ||
| + | |||
| + | |||
| + | ==17/09/2010== | ||
| + | * colony PCR, mini-prep and digestion of promising psiCHECK or TetR constructs containing binding site | ||
| + | ** control plates with horrible number of re-ligations | ||
= 17/09/2010 = | = 17/09/2010 = | ||
| Line 368: | Line 442: | ||
* plenty of colonies on the P56+R8 and P56+R11, NO colonies on control plate with SAPed backbone, colony PCR revealed 50% of positive clones | * plenty of colonies on the P56+R8 and P56+R11, NO colonies on control plate with SAPed backbone, colony PCR revealed 50% of positive clones | ||
<br/> | <br/> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{{:Team:Heidelberg/Pagemiddle}} | {{:Team:Heidelberg/Pagemiddle}} | ||
Revision as of 11:24, 25 October 2010

|
||
 "
"