Lab notes (7/12 - 7/18)
Group: Flagella
Exp. 1
Methods: Miniprep, NanoDrop and gel electrophoresis
protocols: MP1.1
Notes: We extracted the plasmid using 9 ml overnight (ON) culture (Stationary phase cells) and Fermentas GeneJET Plasmid Miniprep kit. We mixed 2 microlitres of the plasmid product and 8 microlitres 1x agarose loading dye and loaded on a 1,5% agarose gel and used Generuler DNA ladder mix (100-10000 nucleotides) from Fermentas as marker.
Results: Nanodrop: Tube 1: 30.7 ng/uL Tube 2: 27.4 ng/uL
Gel electrophoresis: No result
--Louise 15:00, 9 July 2010 (UTC)
Exp. 2
Methods: gel electrophoresis
Notes: We ran another gel electrophoresis on the miniPrep sample form above. We loaded 4 microlitres of sample and 4 microlitres 1x agarose loading dye on a 1,5% agarose gel, used Generuler DNA ladder mix (100-10000 nucleotides) from Fermentas as marker.
Results: Gel electrophoresis: The pSB3K3 plasmid is 2750bp long and the RFP with generator is 1096 bp, which should give a band at about 3850bp. Bands at this size were detected.
Exp. 3
Methods: MiniPrep, NanoDrop and gel electrophoresis.
protocols: MP1.1
Notes: We used 10 ml af a culture in exponential phase (1ml cells from an ON culture + 9ml LB medium, incubated at 37 degrees celcius for 4 hours) and Fermentas GeneJET Plasmid Miniprep kit. We mixed 4 microlitres of sample and 4 microlitres 1x agarose loading dye and loaded on a 1,5% agarose gel and used Generuler DNA ladder mix (100-10000 nucleotides) from Fermentas as marker.
Results: Nanodrop: Tube 1: 38.7 ng/uL Tube 2: 32.4 ng/uL.
Gel electrophoresis: Bands were detected.
--Louise 17:21, 12 July 2010 (UTC)
Amplification of FlhDC, FlhD and FlhC genes
Date: July the 9th
Experiment done by: Sheila and Pernille
Methods: PCR and Gel electrophoresis.
Protocols: CHP1.2
Notes: We decided to test our primers on previously purified cromosomal DNA. Examination of the primers showed that the FlhC reverse primer had a melting temperature of only 45˚C. Therefore we decided to run the samples on a gradient PCR. Simultaneously, we prepared 2 extra samples, isolating FlhD and FlhC, respectively. We did this because we wanted to see if our problems were caused because the combined gene-sequence was to long (932bp).
Because we just wanted to test our primers in this PCR, we used Taq polymerase, because although it doesn’t proofread, it is remarkably cheaper than Pfu. On the [http://www.fermentas.com/en/products/all/pcr-qpcr-rt-pcr/standard-pcr/ep028-taq-dna-native Fermentas homepage] we found that the annealing temperature for Taq is Tm-5 , which in this case means 40˚C. However, Taq polymerase is not very effective at temperatures under 50˚C so we designed the gradient to lies between 40 and 55˚C. The PCR machine chose the following temperatures:
Column
|
Temperature (Celcius)
|
1
|
39.9
|
2
|
40.3
|
3
|
41.2
|
4
|
42.6
|
5
|
44.3
|
6
|
46.3
|
7
|
48.3
|
8
|
50.5
|
9
|
52.1
|
10
|
53.6
|
11
|
54.6
|
12
|
55.1
|
We then chose six temperatures to test:
Sample
|
Temperature (Celcius)
|
1
|
40.3
|
2
|
42.6
|
3
|
44.3
|
4
|
46.3
|
5
|
48.3
|
6
|
50.3
|
The following table shows the Gradient PCR program:
Temperature (Celcius)
|
Time (min)
|
Description
|
94
|
3
|
Initialization |
94
|
2
|
Denaturation |
40-55
|
½
|
Annealing |
72
|
2
|
Elongation
|
-
|
x 29
|
GOTO step 2
|
72
|
5
|
Final Elongation |
4
|
|
Hold |
Results: The experiment was succesfull! We could detect FlhDC DNA at temperatures between 42.6˚C and 48.3˚C. FlhD DNA at temperatures between 40.3˚C and 44.3˚C and also between 48.3˚C and 50.3˚C. FlhC DNA at temperatures run between 40.3˚C and 50.3˚C.
--
Sheila 08:46, 14 July 2010 (UTC)
--
Pernm07 09:41, 14 July 2010 (UTC)
Amplification of FlhDC with Pfu
Date: July 12th
Experiment done by: Pernille
methods: PCR and gel electophoresis
Protocols: CHP1.2
Notes: The purified chromosomal DNA was tested with Pfu; a polymerase with proofreading ability. The DNA used was from the E. coli strain MG1655 (tube 17). The annealing temperature was used in the PCR program was 44˚C. We decide to use this temperature while it is in the middle of the temperature span we successful used in the last experiment. Further we increas the elongation time to 1 min and 30 sec. Other than these changes we followed the PRC program decribed in the protocol. The PCR product was run on a 1,5% agarose gel.
results: Unfortunately the experiment did not give any results on the gel.
--Pernm07 10:56, 13 July 2010 (UTC)
--Sheila 08:47, 14 July 2010 (UTC)
Amplification of FlhDC with pfx
Date: July 13th
Experiment done by: Ann and Pernille
methods: The experiment was perform in the E.coli strain MG1655. The DNA purification was done in duplicates and are saved in the freezer in tube 8 and 9 (green label). The DNA concentration was measured by NanoDrop and the concentrations are 16,87ng/uL and 19,25ng/uL, respectively.
protocol: GP1.1 and CP1.1.
Notes: The experiment was generally performed corresponding to the protocol but small modifications was made. In the DNA purification experiment we used 300 microlitres instead of 200 microlitres of the overnight culture and it is important to remember that the 800 microlitres precipitation solution, containing 80 microlitres solution and 720 microlitres deionized water, are made directly before use.
Because our PRC for the time being haven't been successful we are trying to do PCR with pfx which is another polymerase with proofreading ability. The PRC program used for running PCR with pfx is as described below:
1
|
start
|
94°C
|
2 min
|
2
|
Denaturing
|
94°C |
1min
|
3
|
Annealing
|
48°C |
1 min
|
4
|
Elongation
|
68°C |
2 min
|
5
|
GOTO 2
|
|
Rep x 29
|
6
|
End
|
68°C |
3min
|
7
|
Hold
|
4°C |
|
Results: No lanes appeared when we ran the gel electophoresis. Therefore the next step is to run PCR with Taq polymerase to check if the problem is the pfu polymerase or if the DNA has not been purifyed properly.
--
Pernm07 10:19, 13 July 2010 (UTC)
--
Sheila 08:49, 14 July 2010 (UTC)
Digestion of FlhD/C with pstI
Exp. 1
Methods: digestion and gel electrophoresis
protocols: RD1.1
Notes:
To verify the restriction site at 822bp in the FlhDC gene, FlhDC PCR product from Taq PCR was digested with pstI restrction enzyme. Samples were loaded onto a 2% agarose gel. Generuler 100bp DNA ladder was used as marker. Undigested FlhDC PCR product was used as control.
Results:
Gel electrophoresis:
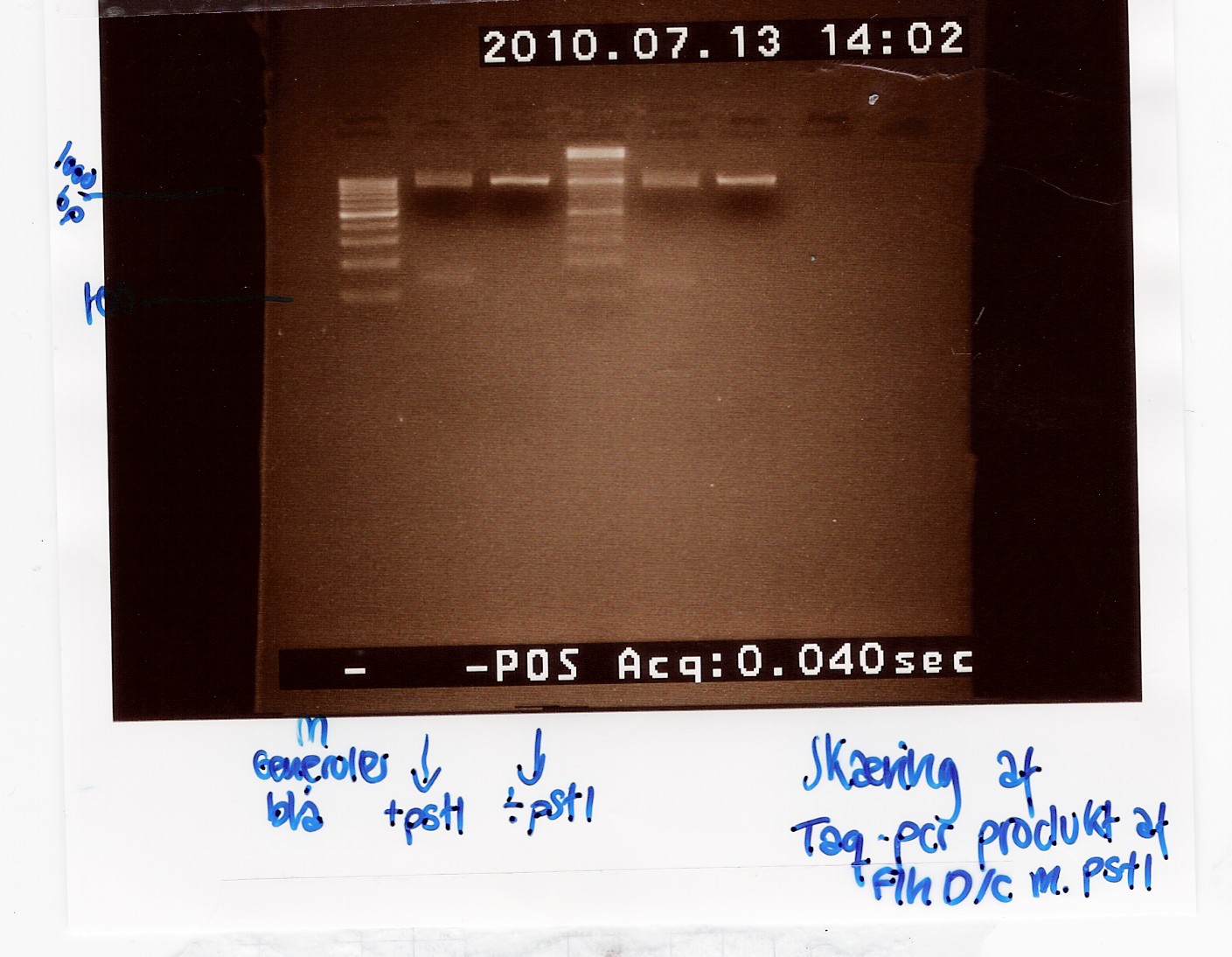
Analysis:
Lane 2 and 5 contains the samples cut with pstI and shows a small DNA fragment below 100 bp. This seems to be primer dimers since the uncut bands lie at the same position as the cut. The experiment has to be made again.
--Tipi 16:44, 19 July 2010 (UTC)
Amplification of FlhDC with Taq
Date: July 14th
Experiment done by: Pernille
Methods: The experiment was performed in the E.coli strain MG1655, from freezer tube 8 and 9 wich was also used for amplification of FlhDC with pfx. The experiment was preformed according to the CP1.1 protocol.
Protocol: CP1.1.
Results: we did not get any results
--Pernm07 07:51, 14 July 2010 (UTC)
--Sheila 09:08, 14 July 2010 (UTC)
Amplification of pSB3K3 with Taq
Start date: July 14th 2010
Methods: The purified pSB3K3 plasmids were amplified by PCR using Taq, as a test, before using the more expensive Pfu.
Protocol: CP1.2
Notes: The plasmids were collected from tubes green 10 and green 11 in the freezer. VR and VF2 primers were used.
The experiment were performed simultaneously with the Taq control version of the extraction of B0034 from pSB1A2 plasmid done by the phototaxis goup
--Sheila 09:08, 14 July 2010 (UTC)
Genomic DNA purification
Date: July 14th
Experiment done by: Pernille and Louise
Methods: We used a overnight culture of E. coli MG1655 cells
Protocols: GP1.1
Notes: We made four tubes with 300 microlitres of E. coli MG1655 cell media instead of 200 microlitres. In tube 1 and 2 we unfortunately added 800 microlitres un-diluted precipitation solution, in tube 3 and 4 we diluted the precipitation solution according to the GP1.1 protocol.
Results: The concentration of the purified DNA was measured by NanoDrop. Tube 1 and 2 did not contain any DNA which we had expected because of the high concentration of precipitation soultion. The DNA concentration in tube 3 and 4 were 28,38ng/uL and 19,4ng/uL, respectively. Tube 3 and 4 were afterwards pooled and saved in the freezer (labeled with white tag 1). The purified DNA was run on a agarose gel to check if it actual contains DNA.
--Louise 10:44, 14 July 2010 (UTC)
--Pernm07 09:33, 14 July 2010 (UTC)
Genomic DNA purification
Date: July 15th
Experiment done by: Sheila
Methods: The DNA purification was performed on an overnight culture of the E. coli strain MG1655 . Three samples of 400 microlitres of culture were extracted, centrifuged, the supernatant removed and the pellet was redissolved in 200 microlitres water
Protocol: GP1.1
Notes:
Results: Not a lot of DNA was detected with the NanoDrop. The samples yielded concentrations of 9,88ng/µL, 13,5ng/µL and 5,3ng/µL respectively. The gel showed 4 bands of DNA in each sample. The samples were located at 200bp, 950bp, 1500bp and >10000bp. The bands less visible in lane 3, which were loaded with sample 3, which also yielded the least concentration.
--Sheila 09:20, 15 July 2010 (UTC)
Amplification of FlhDC regulon from purified cromosomal DNA using TAQ
Experiment done by: Christian
Date: July 14th
Methods: PCR on purified chromosomal DNA
Protocols: CP1.2
Notes: 4 tubes loaded with DNA from freezer: 2x(8 green) and 2x(9 green). Both tubes were discarded as they were empty.
(8 Green) was dilluted 4x to make pipetting possible.
Primers used were FlhD fw and FlhC rw.
Anealing temperature was set to 48°C and elongation was set to 2 minutes.
1,5% agarose gel was loaded with Generuler DNA ladder mix.
Results:Band showed at around 3kb in both wells for (8 green) and one well for (9 green)
Analysis: The FlhDC regulon was not extracted.
--CKurtzhals 16:11, 15 July 2010 (UTC)
Checking the new primers
Experiment done by: Pernille and Louise
Date: July 15th and 16th
Protocols: CP1.2
Note: Polymerase used: TAQ
Methods: PCR on purified E. coli MG1655 chromosomal DNA from Gel electrophoresis.
We used the four new primers we designed in the order: FlhDC fw, FlhDC rev, FlhDCmut fw and FlhDCmut rev.
We ran a gradient PCR with six different annealing temperatures for three primer combinations (Tables below).
PCR-mixes:
PCR-mix 1
|
PCR-mix 2
|
PCR-mix 3
|
| 36 ul Water |
36 ul Water |
36 ul Water |
| 5 ul 10x TAQ Buffer |
5 ul 10x TAQ Buffer |
5 ul 10x TAQ Buffer |
| 1 ul 10 mM dNTP |
1 ul 10 mM dNTP |
1 ul 10 mM dNTP |
| 2 ul MgCl2 |
2 ul MgCl2 |
2 ul MgCl2 |
| 2 ul FlhDC fw Primer (Freeze
tupe white 5) |
2 ul FlhDCmut fw Primer (Freeze
tupe white 7)
|
2 ul FlhDC fw Primer (Freeze
tupe white 5)
|
| 2 ul FlhDCmut rev Primer (Freeze
tupe white 6) |
2 ul FlhDC rev Primer (freeze
tupe white 4)
|
2 ul FlhDC rev Primer (Freeze
tupe white 4)
|
| 1 ul Purified chromosomal DNA
from MG1655 (Freeze tupe white 9) |
1 ul Purified chromosomal DNA
from MG1655 (Freeze tupe white 9) |
1 ul Purified chromosomal DNA
from MG1655 (Freeze tupe white 9) |
| 1 ul TAQ (added just before PCR
start)
|
1 ul TAQ (added just before PCR
start) |
1 ul TAQ (added just before PCR
start) |
PCR Program:
Time:
|
Temperature (Celcius):
|
Process:
|
3 min
|
94
|
Start
|
2 min
|
94
|
Denaturation
|
30 sec
|
Gradient
|
Annealing
|
2 min
|
72
|
Elongation
|
x 29
|
-
|
GOTO step 2
|
5 min
|
72
|
End
|
DONE!
|
4
|
Hold
|
Annealing Gradient:
Column
|
Temperature (Celcius)
|
4
|
48.5
|
5
|
50.8
|
6
|
53.4
|
7
|
56.1
|
9
|
60.1
|
11
|
64.5
|
Gel electrophoresis
Method: We used a 1.5% agarose gel and two ladders (GeneRuler DNA ladder mix), 8 microlitres PCR product was loaded together with 2 microlitres agarose loading dye
Loading Table
Well No.
|
PCR Loading
|
Primers
|
1
|
GeneRuler DNA ladder mix
|
-
|
2
|
3.4 (PCR-mix 3. Column 4)
|
FlhDC fw + FlhDC rev
|
3
|
3.5
|
----II----
|
4
|
3.6
|
----II---- |
5
|
3.7
|
----II---- |
6
|
3.9
|
----II---- |
7
|
3.11
|
----II---- |
8
|
2.4
|
FlhDCmut fw + FlhDC rev
|
9
|
2.5
|
----II---- |
10
|
2.6
|
----II---- |
11
|
2.7
|
----II---- |
12
|
2.9
|
----II---- |
13
|
2.11
|
----II---- |
14
|
1.4
|
FlhDC fw + FlhDCmut rev
|
15
|
1.5
|
----II---- |
16
|
1.6
|
----II---- |
17
|
1.7
|
----II---- |
18
|
1.9
|
----II---- |
19
|
1.11
|
----II---- |
20
|
GeneRuler DNA ladder mix
|
-
|
Result:
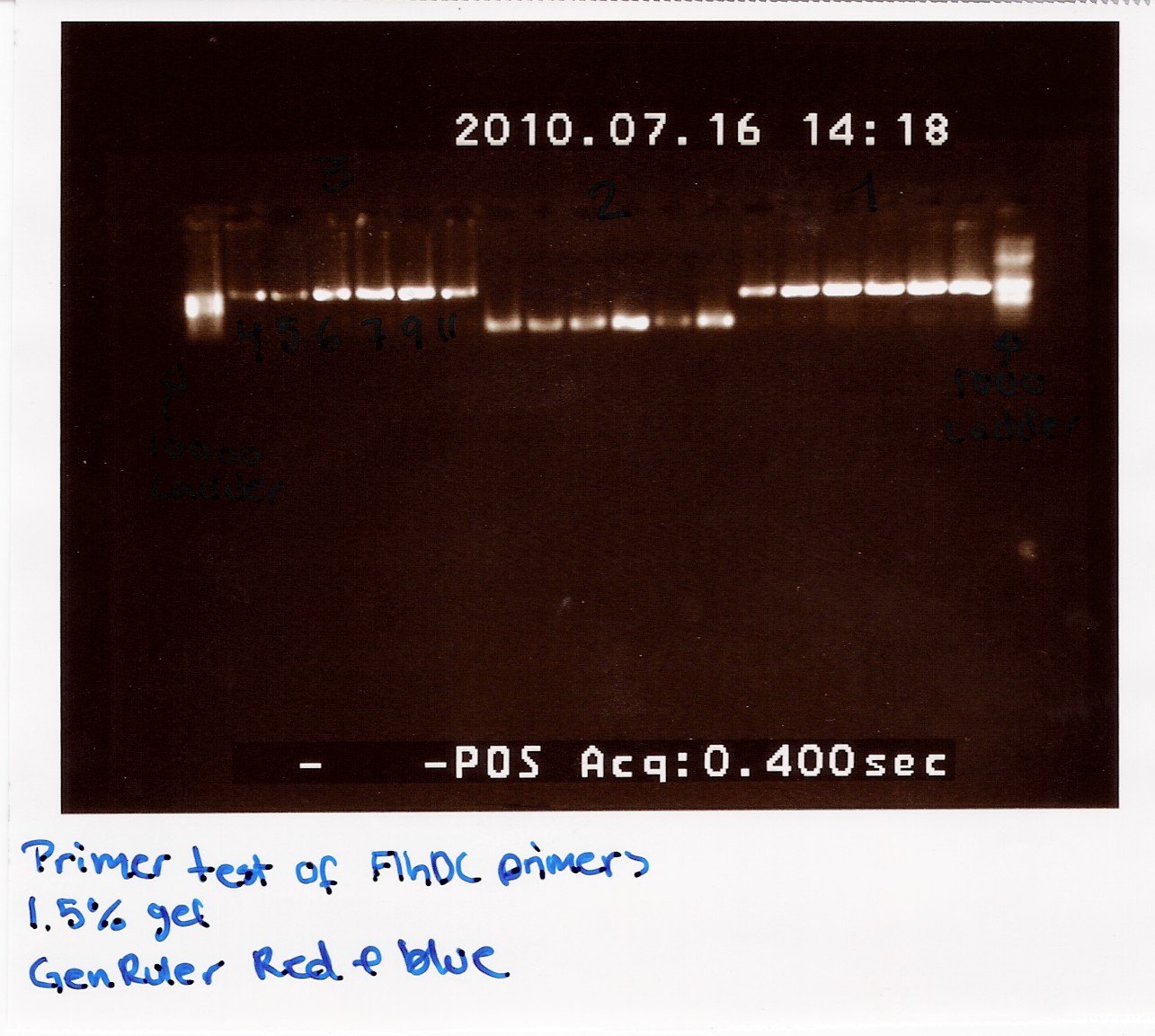
The image shows PCR results with the right lengths. Strong expression for PCR with primers; FlhDC fw and FlhDCmut rev is seen at temperatures 53.4 Celcius, 56.1 Celcius and 60.1 Celcius, FlhDCmut fv and FlhDC rev is seen at 56.1 celcius, and FlhDC fw and FlhDC rev is seen at all gradient temperatures. The Gradient shows a good result for all three PCR's at 56.1 Celcius.
--Louise 12:50, 16 July 2010 (UTC)
Amplification of FlhDC with Pfu and mutation of restrictionsite, located in FlhC
Experiment done by: Sheila
Date: July 16th
Protocols: CP1.1
Note: Polymerase used: Pfu. Three different samples were prepared as was done,when the new primers were tested with Taq (one, copied the entire operon, the second copied the gene from FlhD to the restriction site in FlhC and the third copied from the restriction site to the end of the FlhC gene), and run at three different temperatures: 50,8˚C, 56,1˚C and 64,5˚C respectively. The temperatures were chosen based on the clearest bands in the gel run in the before mentioned experiment.
Methods: PCR on purified E. coli MG1655 chromosomal DNA from Gel electrophoresis.
Primers used: FlhDC fw, FlhDC rev, FlhDCmut fw and FlhDCmut rev.
Results: At last, we have PCR results. The gel showed clear bands of both mutated pieces of the gene, as well as the entire operon. And because this PCR was run with Pfu, we can actually use these results and move on with the project.
--Sheila 15:31, 16 July 2010 (UTC)
Group: Photosensor
No experiments have been caried out this week. We are waiting for the DNA to arrive.
Group: Retinal
Transformation of TOP10 e. coli with [http://partsregistry.org/Part:BBa_K274210 K274210]
Experiment continued from last week.
Colony PCR of K274210 in pSB1A2
Start date: July 12th
Experiment done by: Christian, Lars Christian
Methods: Colony PCR (modified)
Protocol: CP1.2
Notes: MgCl2 was added as a gradient, samples 1-2 contain 2 microlitres (0,04255), samples 3-5 2,25 microlitres (0,04787). We mistakenly added 30µl (instead of 17µl) premix to samples 6 - 9, so the concentration of MgCl2 is reduced. Sample 6 contains 2,25 microlitres (0,0375), samples 7-8 2,5 microlitres (0,04167), sample 9 2,75 microlitres (0,04583).
Gel was loaded with DNA-ladder plus, with the upper marker at 5000 bp.
Results: We found plasmids at the expected 5 kb marker in colonies 1-8. Successfull plates were kept in incubator over night. They will be made into overnight cultures and thereafter frozen.
Analysis: It seems we have transformed our cells with K274210. Confirmation will come when we run experiments to demonstrate presence of beta-carotene.
Transformation of [http://partsregistry.org/Part:BBa_R0011 R0011 ]and [http://partsregistry.org/Part:BBa_I0500 I0500]
Start date: July 12th End date:July 14th
Methods: Overnight culture, competent cells, transformation.
Protocols:CC1.1, TR1.1.
Making competent E.Coli TOP 10
Experiment done by: Christian, Maria and LC
Date: July 12th
Notes: Everything was done according to protocol.
Results: We made competent cells for use within 48 hours.
Analysis:
Transformation of pBad and lacl promoter
Experiment done by: Christian, Maria and LC
Date: July 12th - 13th
Notes: Added 200 microlitres of the un-pelleted cells, instead of 150µl to the agar plates.
Some of the agar dishes were damaged during drying and plating. We tried to use them anyway.
Results: No plates showed colonies, apart from the positive control
Analysis: We suspect the agar dishes might have been the cause of this.
Mini-prep of pSB1A2 w. [http://partsregistry.org/Part:BBa_B0034 B0034], pSB1AK3 w. [http://partsregistry.org/Part:BBa_B0015 B0015] and pSB3K3 w. [http://partsregistry.org/Part:BBa_J04450 J04450] (transformation from 08/07)
Start date: July 12th
Methods: Mini-prep
Protocol: MP1.2
Notes:
Samples were loaded into a 1% agarose gel with GeneRuler DNA ladder mix from Fermentas as marker.
Results:
Gel electrophoresis:
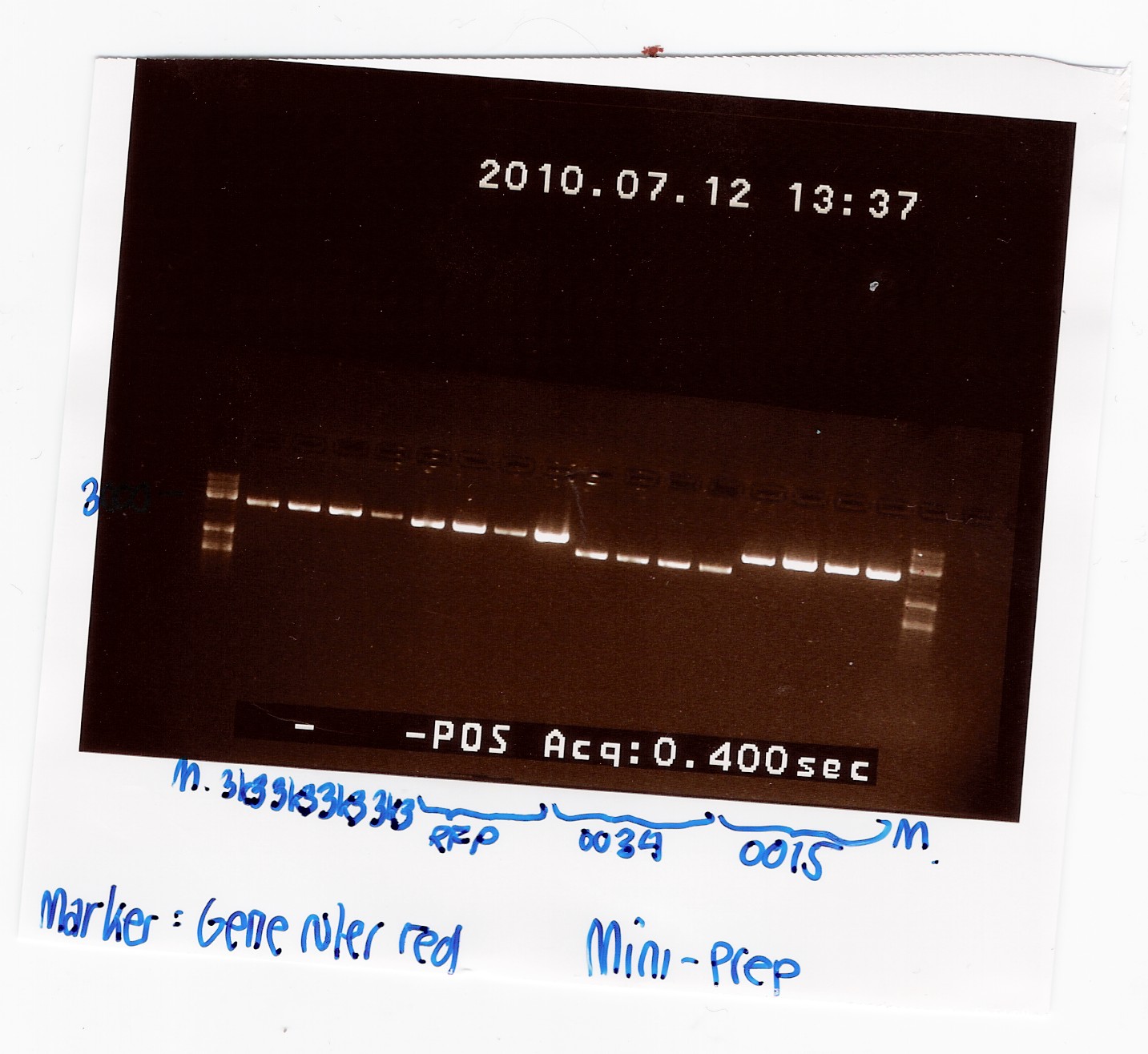
Nanodrop:
Sample ID
|
ng/uL
|
260/280
|
260/230
|
pSB1K3 1
|
41.09
|
1.88
|
1.93
|
pSB1K3 2
|
44.44
|
1.83
|
1.7
|
pSB1K3 3
|
45.37
|
1.9
|
1.69
|
pSB1K3 4
|
43.87
|
1.91
|
1.88
|
RFP 1
|
85.11
|
1.88
|
2.05
|
RFP 2
|
95.32
|
1.86
|
2.07
|
RFP 3
|
87.75
|
1.9
|
2.13
|
RFP 4
|
98.04
|
1.87
|
1.92
|
B0034 1
|
59.33
|
1.88
|
2.00
|
B0034 2
|
59.73
|
1.88
|
2.06
|
B0034 3
|
67.95
|
1.9
|
2.12
|
B0034 4
|
72.03
|
1.87
|
2.17
|
B0015 1
|
89.29
|
1.9
|
2.25
|
B0015 2
|
101.76
|
1.88
|
2.24
|
B0015 3
|
84.33
|
1.89
|
2.09
|
B0015 4
|
88.59
|
1.87
|
2.2
|
Analysis:
A clear band was seen in all lanes, so we have succesfully isolated DNA.
All bands appeared to be about 1000 kb smaller than the expected size. This could be due to the supercoiling of the plasmids. To verify this, the plasmids were cut with PstI: Digestion of plasmids
Digestion of pSB1A2 w. [http://partsregistry.org/Part:BBa_B0034 B0034], pSB1AK3 w. [http://partsregistry.org/Part:BBa_B0015 B0015] and pSB3K3 w. [http://partsregistry.org/Part:BBa_J04450 J04450]and pSB1A2 using pstI(miniprep from 12/07)
Start date: July 13th
Methods: Digestion
Protocol: RD1.1
Experiment done by: Maria and Sheila
Notes: Digestion of the plasmids with a single restriction enzyme was done to ensure that the plasmids extracted from mini-prep has the correct size. Digestion was done with half the amount of restriction mixture.
Samples were loaded into a 1.5% agarose gel with GeneRuler DNA ladder mix from Fermentas as marker.
Results:
Gel electrophoresis:
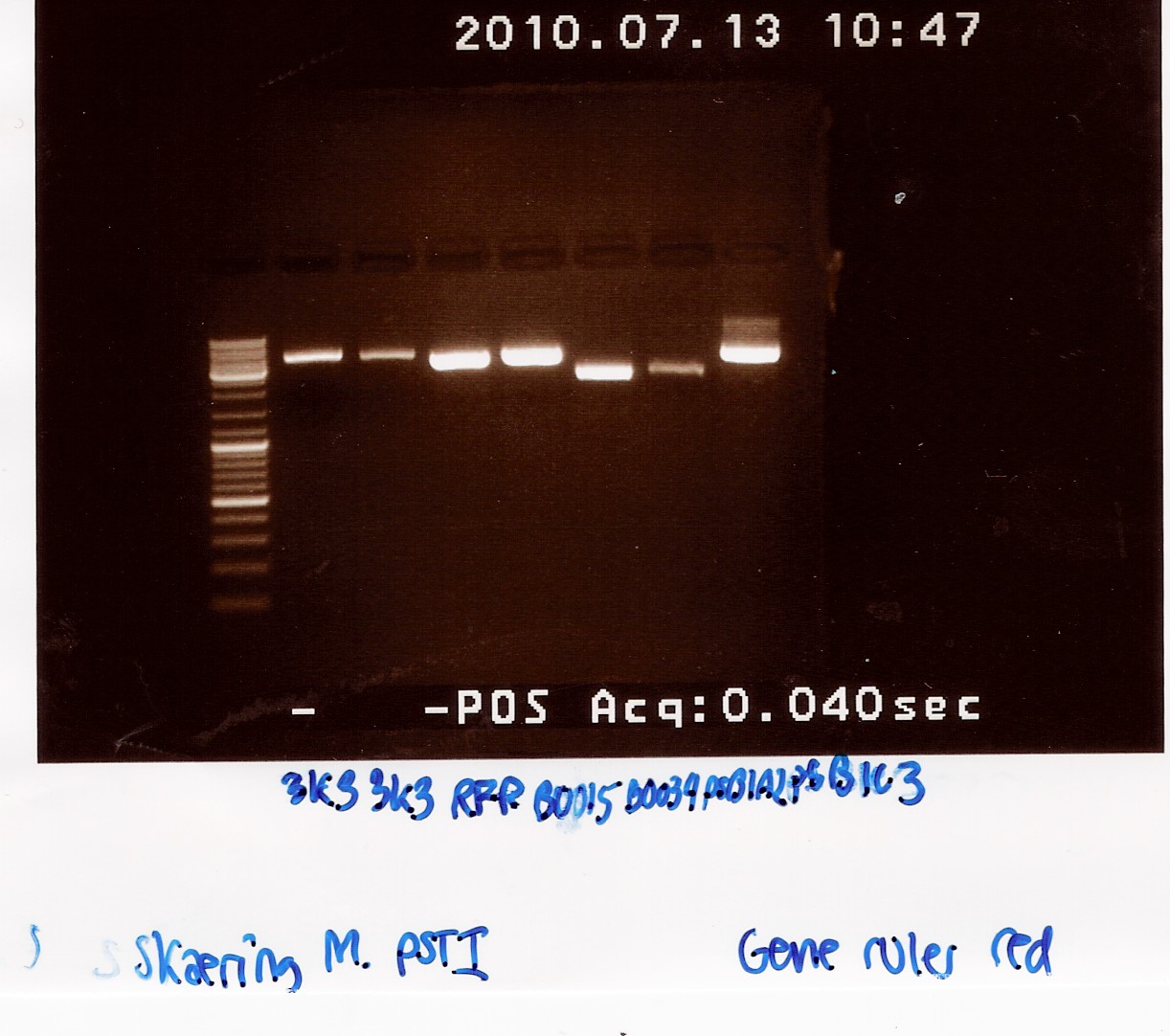
Analysis:
All bands had the size we expected for the plasmids. We therefore expect that we have isolated the right plasmids.
The samples from miniprep 12/7 was pooled and stored as no. 24, 25, 26 and 27 at -20 degrees C.
--Tipi 15:53, 19 July 2010 (UTC)
Transformation of E. coli MG1655 with [http://partsregistry.org/Part:BBa_K274210 K274210], [http://partsregistry.org/Part:BBa_E0040 E0040] and [http://partsregistry.org/Part:BBa_I0500 I0500]
Start Date: July 13th
Methods: overnight cultures, Competent Cells, Transformation of competent cells.
Protocols: CC1.1, TR1.1
Experiment: Making competent E. coli MG1655
Experiment done by: Christian
Date: July 13th
Notes: Growth was started at 09.30 with an OD550 of 0.19A. The cells were harvested at 10.26 where OD550 was 0.48.
Results: Competent cells were made according to protocol. Apparently E. coli MG1655 reaches OD550 0.5 at least an hour faster than E. coli TOP10.
Experiment: Transforming E.coli MG1655 strain with E0040 (GFP coding sequence) and I0500 (pBad arabinose inducible promoter)
Date: July 13th
Notes:
I0500 is located in pSB2K3. E0040 is in pSB1A2. 200 microlitres cells were added to each tube. Positive control is RFP generator as usual. Negative control is just cells.
Only 1 microliter DNA material was transferred per tube, since it was taken from the iGEM 2010 distribution plates.
Plates were dried at 42°C for 15 minutes, just prior to plating. Upon plating we discovered a problem with many of the ampicilin plates we made a couple of days ago. The agar breaks on the slightest contact, and are therfore impossible to use.
As a result of the defective agar plates, most of our cells carrying E0040 were ruined. We managed to save and plate out two batches, one from each tube, so we are hoping for the best tomorrow.
Results: Only cells with E0040 grew colonies by overnight incubation.
Miniprep of [http://partsregistry.org/Part:BBa_K098995 K098995] in pSB1A2
Start date: July 13th
Methods: Fermentas GeneJET plasmid miniprep kit
Protocol: MP1.1
Experiment done by: LC
Notes: Centrifuged the overnight culture for 15 mins. at 3000 rpm. Used 10 ml of culture in each tube and added 500 microlitres resuspension solution, then split it into two samples.
Results:
Results were as expected (correct), the plasmid was around 3000 bp long on the gel.
Analysis: All four samples turned out fine, so they were pooled and frozen. (See results for details)
Nr. 28 in the freezer = K098995
Preliminary PCR to amplify VF2-VR piece containing part and restriction sites
Date: July 14th
Methods: PCR of DNA in solutions
Protocol: CP1.1-PFU protocol
Experiment done by: Christian
Notes: DNA material from tube 27 was taken from the freezer. Primers were standard VF2 and VR of which we used 10 micromolars. PFU buffer with MgSO4 was used. No further MgSO4 was added. A TAQ control was done in a parallel experiment.
Protocol was followed to point, with the exception of 1 microliter additional H2O being added because of an error. It will yield 51 microlitres in each pcr reaction.
Program was run according to protocol. Elongation time was set to 1 minute and 30 seconds.
A control on the same DNA was run using TAQ polymerase. It was run on the same protocol and program as a parallel experiment in the flagella group: Amplification of pSB3K3 with taq
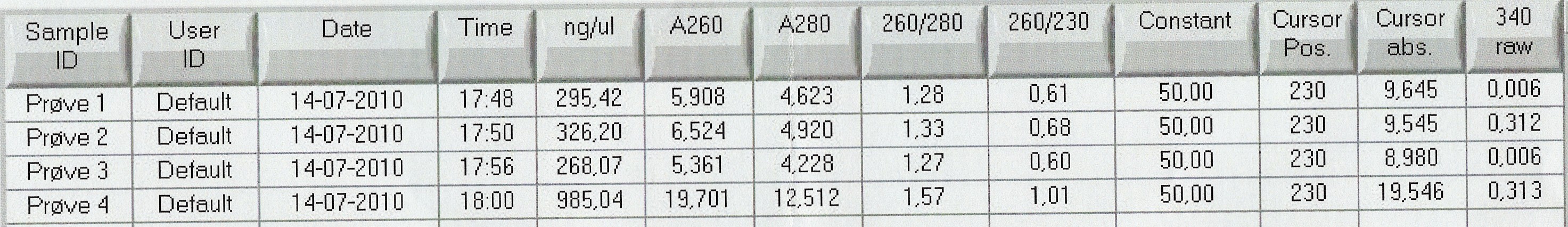
Results: Bands showed in all four wells on the gel, located at the expected length of 250 bp. Nanodrop showed some contamination, but a good DNA concentration of 345ng/uL. The contamination might be the entire plasmid, since elongation time was a little in the high end.
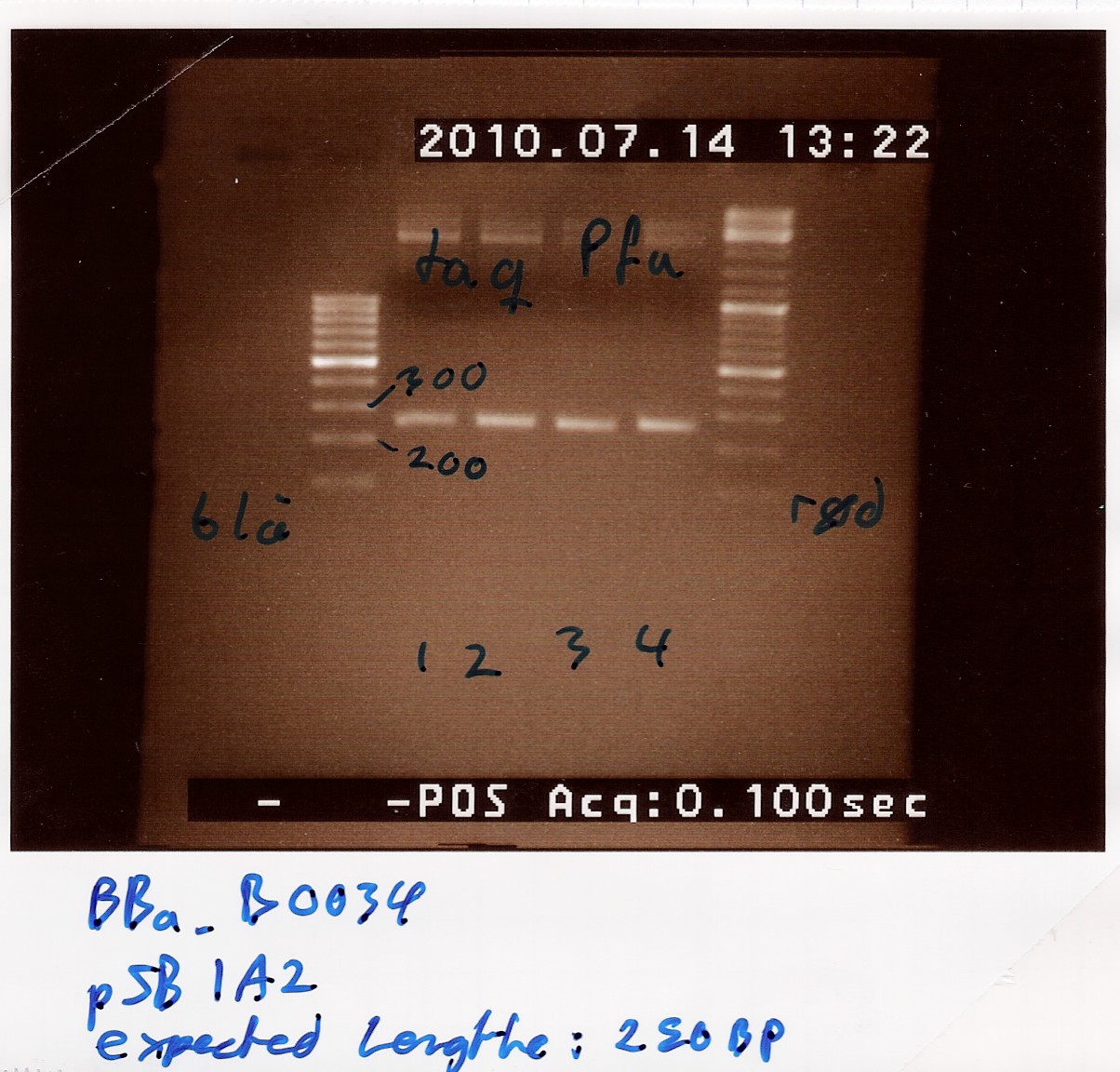
Analysis: The PCR product might be to contaminated still, and there's too little anyway. Therefore The PCR product will need to be further amplified.
Further amplification of PCR product
Date: July 14th
Methods: PCR of DNA in solutions
Protocol: CP1.1-PFU protocol
Experiment done by: Christian
Notes: 4 tubes were loaded, using one microliter of DNA template from the above experiment. Protocol was followed to point.
The same PCR program was used (PF1 on machine 2), elongation time once again set to 1 minute and 30 seconds.
Remaining PCR product was frozen as (White-2).
A 1% agarose gel was loaded with 5 microlitres samples of each tube and GeneRuler 100bp ladder from Fermentas as marker.
Results: Bands showed at 250bp for all tubes. Tubes 1-3 showed a strange band that was larger than the ladder could show. Tube 4 was free of this band, and also showed a clearer band at 250bp.
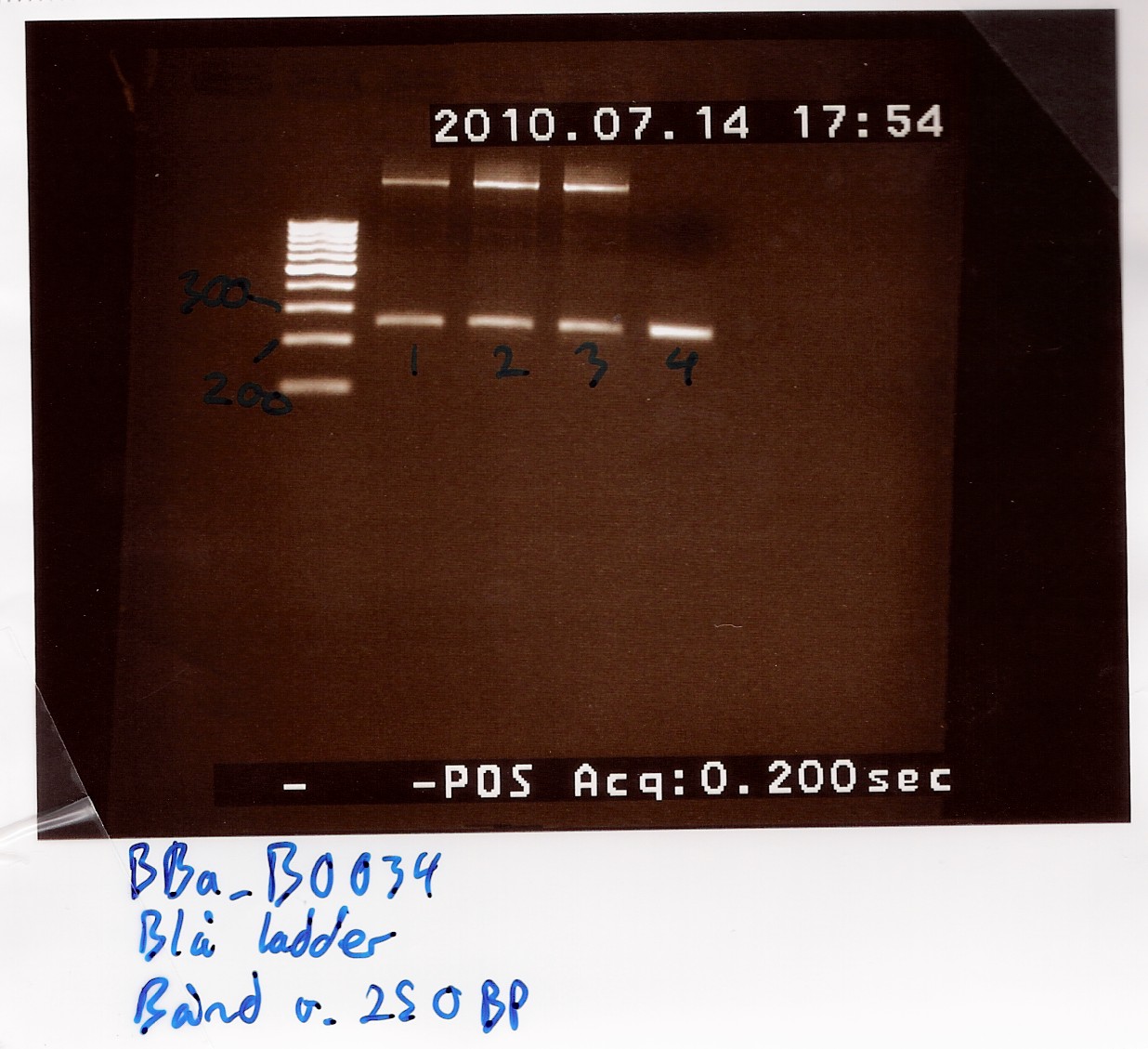
Nanodrop showed signatures for tubes 1-3 like in the samples of the above experiment. DNA was twice as concentrated in tube 4.
--CKurtzhals 16:05, 14 July 2010 (UTC)
Date: July 15th
Methods: Gel Extraction Kit
Protocol: DE1.1
Experiment done by: Christian, Lars Christian
Notes: Four 50 microliter wells were created in the gel to accomodate all the genetic material, which was loaded along with GeneRuler 100bp ladder from Fermentas as marker. We extracted DNA from the 250bp position according to protocol.
After dissolving the gel slice, we added 100% isopropanol to solution in 1:2 ratio, as noted in the kit protocol for DNA pieces less than 500bp.
Lars Christian eluded twice with 30ul elution buffer instead of once with 50ul for 4 of the eight tubes. Otherwise protcol was followed.
Extractions were pooled and frozen as (White 11)
Results: Nanodrop showed 5,00ng/uL DNA with 260/280 ratio at 2.29 and 260/230 ratio at 0.02.
Analysis: This product is not optimal. We will need to decide whether to try again.
--CKurtzhals 16:07, 15 July 2010 (UTC)
Miniprep and transformation of [http://partsregistry.org/Part:BBa_K274210 K274210] in pSB1A2
Start date: July 14.th
Methods: Fermentas GeneJET plasmid miniprep kit
Protocol: MP1.1
Experiment done by: LC
Notes: No pellet after step 4, but still continued. Later we got a small pellet at step 6.
Results:
The miniprep failed as expected, since there was no pellet after the second centrifugation.
Analysis:The experiment will be repeated in the afternoon.
Second try:
Methods and protocol as in the first try.
Notes: We only had 3,5 ml of overnight culture for the miniprep, so we had to manage with that.
Results: Insert gel picture and nanodrop table.
Analysis: The second try worked, so we went on to make transformations with the plasmid. It is in the freezer as nr. 3 (white).
Transformation of the miniprepped plasmid:
Experiment done by: LC, Maria
Protocol: TR 1.1
Notes: Transformed K274210 in pSB1A2 on LA plates with 100 microgram/ml ampicillin into E. coli MG1655. We used 2 microliter from the miniprep product for each sample.
Our suspicion that the agar plates we made are faulty got confirmed, three more plates were destroyed while spreading the bacteria on them (Sample 1a, 2b, 2pellet and the positive control)
Results: Cells grew just fine and the negative control was as expected. After a night at 37° the cells seemed a little more yellowish to the eye, than the usual.
Analysis: The transformations succeeded.
Coloni PCR of pSB1A2 w. [http://partsregistry.org/Part:BBa_E0040 E0040] (transformation 13/7)
Start date: July 14th
Methods: Taq coloni PCR and gel electrophoresis
Protocol: CP1.2
Experiment done by: Maria
Notes:10 colonies from the Transformation 13/7 by taq PCR using 10 micromolar of VF2 and VR.
In PCR tube no. 6 most of the premix was lost.
Results:
Gel electrophoresis:
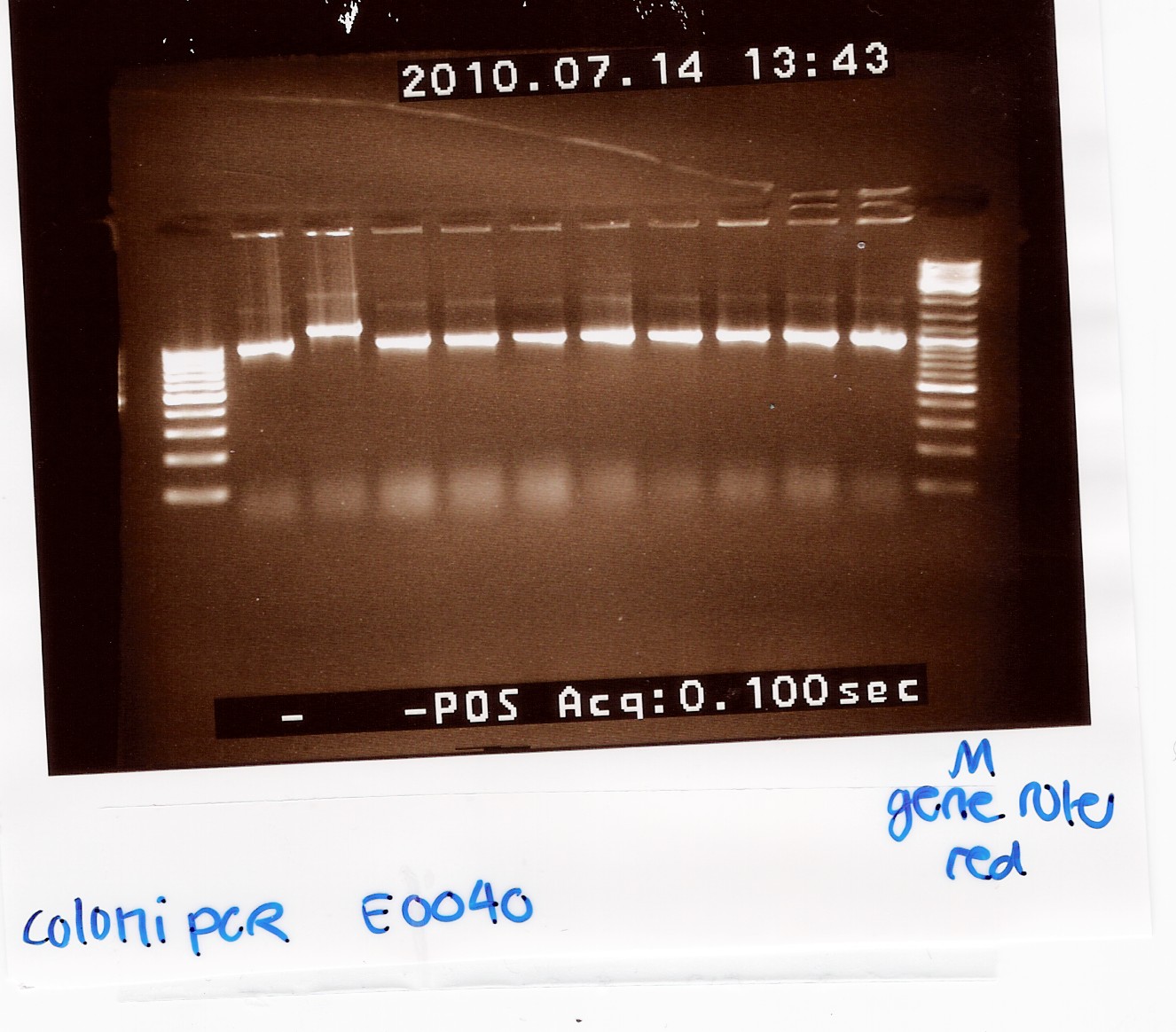
Analysis:
9 of the 10 colonies contained a plasmid of the correct size. plasmid was miniprep'ed from coloni no.10 (miniprep)
--Tipi 16:09, 19 July 2010 (UTC)
Transformation of [http://partsregistry.org/Part:BBa_K081005 K081005] in pSB1A2 (constitutive promoter and RBS combined) and [http://partsregistry.org/Part:BBa_R0011 R0011] in pSB1A2 in Top 10 E.Coli
Start date: July 15th - 16th
Methods: Overnight culture, making competent cells, transformation
Protocol: TR1.1
Experiment done by: LC
Notes:Overnight culture was made of 110 ml LB medium and a frozen top 10 culture. The cells took quite a while to reach an OD550 > 0,5, but it was consistent with our prior experience with E.coli TOP10:
Time
|
Optical density
|
9:28
|
0,02
|
10:36
|
0,029
|
11:07
|
0,076
|
11:37
|
0,185
|
12:07
|
0,288
|
12:50
|
0,52
|
Transformation went OK, only one plate got destroyed this time around.
Results:Both transformations failed, even though positive and negative control were correct.
Analysis:We are unsure as to the reason of the experiment failing, on monday we will try to transform the same brick on different plates to check if it is in the right plasmid (
transformation).
--
Lclund 10:00, 19 July 2010 (UTC)
Miniprep of [http://partsregistry.org/Part:BBa_E0040 BBa_E0040]
Start date: July 16th
Methods: Overnight culture, miniprep
Protocol: MP1.1
Experiment done by: LC
Notes: Followed the modified protocol. (10 ml/falcon tube, 500 ul resuspension solution, then split it into two samples)
Results: 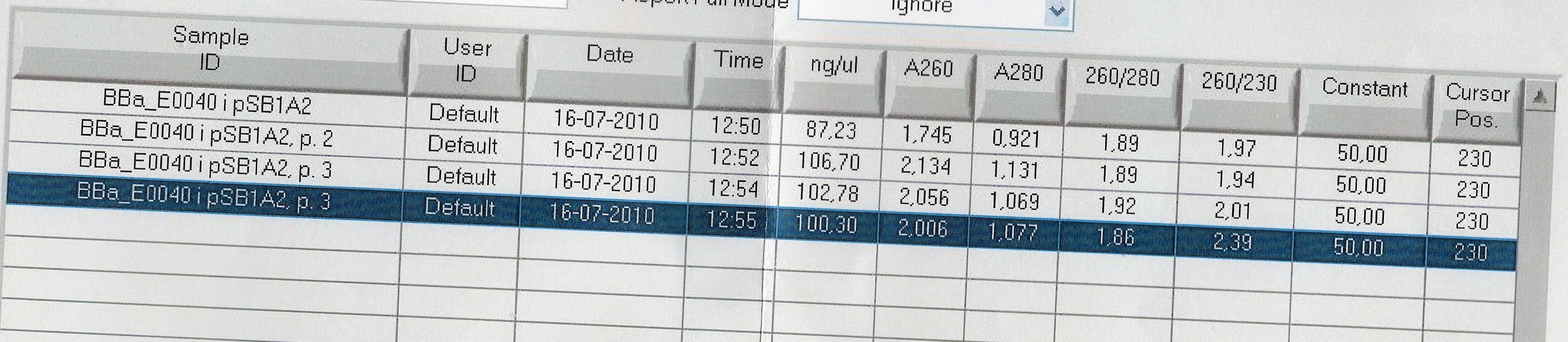
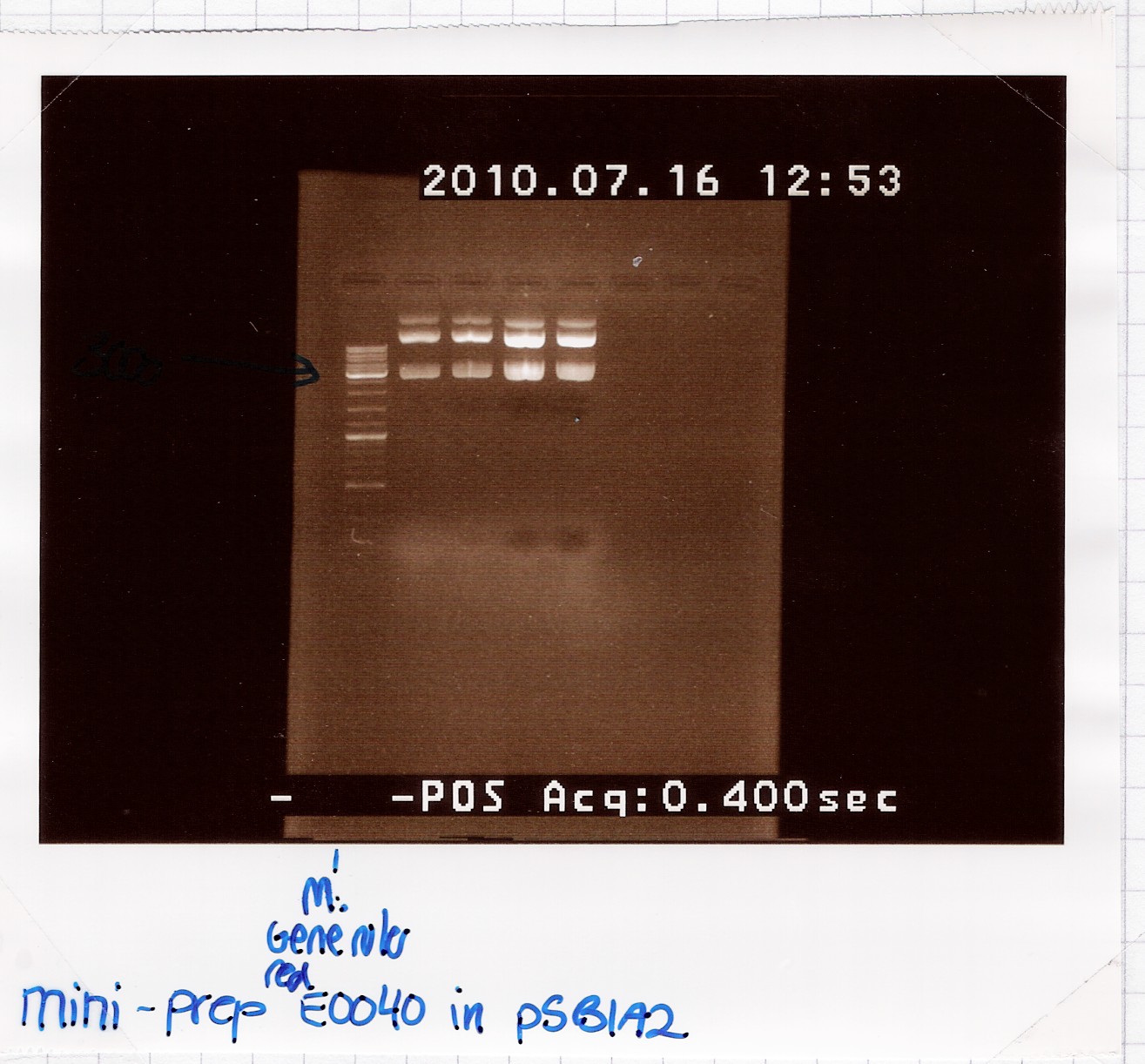
Analysis:Because of the multiple bands we will try cutting with restriction enzymes, to see if the big bands are just supercoiled plasmid.
--Lclund 10:00, 19 July 2010 (UTC)
Restriction digest of [http://partsregistry.org/Part:BBa_E0040 E0040] miniprep product
Start date: July 16th
Methods: Restriction digest
Protocol: RD1.1
Experiment done by: Maria, LC
Notes: /
Results:
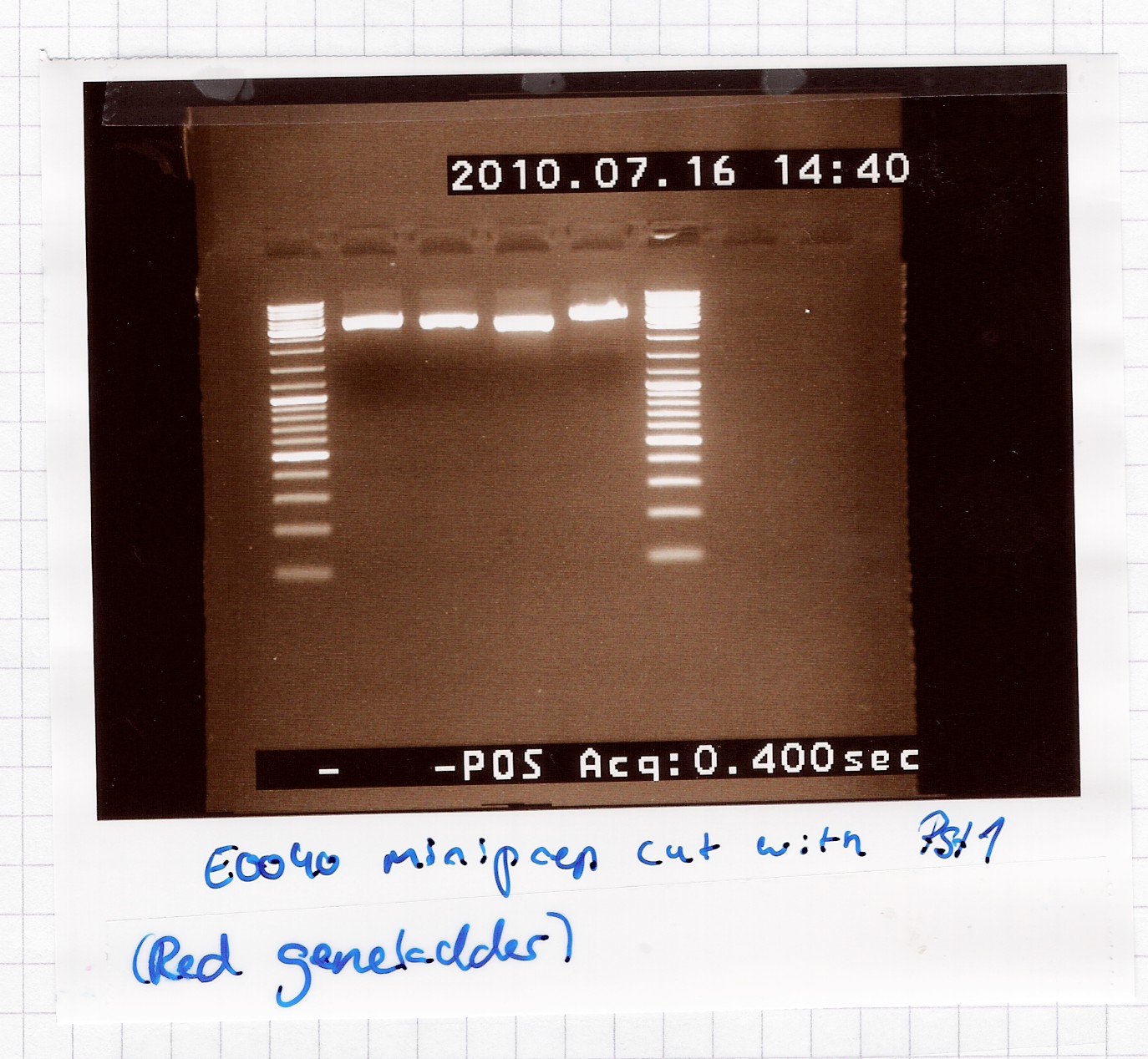
A single band is seen on the gel, which means that the multiple bands in the miniprep gel were only because of supercoiling.
Analysis: Sample 2 and 3 got pooled and frozen, 1 and 4 were discarded, since their length didn't seem exactly right.
Nr. 12 in the freezer = E0040 miniprep product in pSB1A2.
--Lclund 10:00, 19 July 2010 (UTC)
 "
"










