Team:Uppsala-SwedenWeek10
From 2010.igem.org
(New page: {{Template:Uppsala}}<!--Do not remove the first and last lines in this page!--> == Week-10 ==) |
(→Week-10) |
||
| Line 1: | Line 1: | ||
{{Template:Uppsala}}<!--Do not remove the first and last lines in this page!--> | {{Template:Uppsala}}<!--Do not remove the first and last lines in this page!--> | ||
== Week-10 == | == Week-10 == | ||
| + | |||
| + | == Construction of G1- G7 == | ||
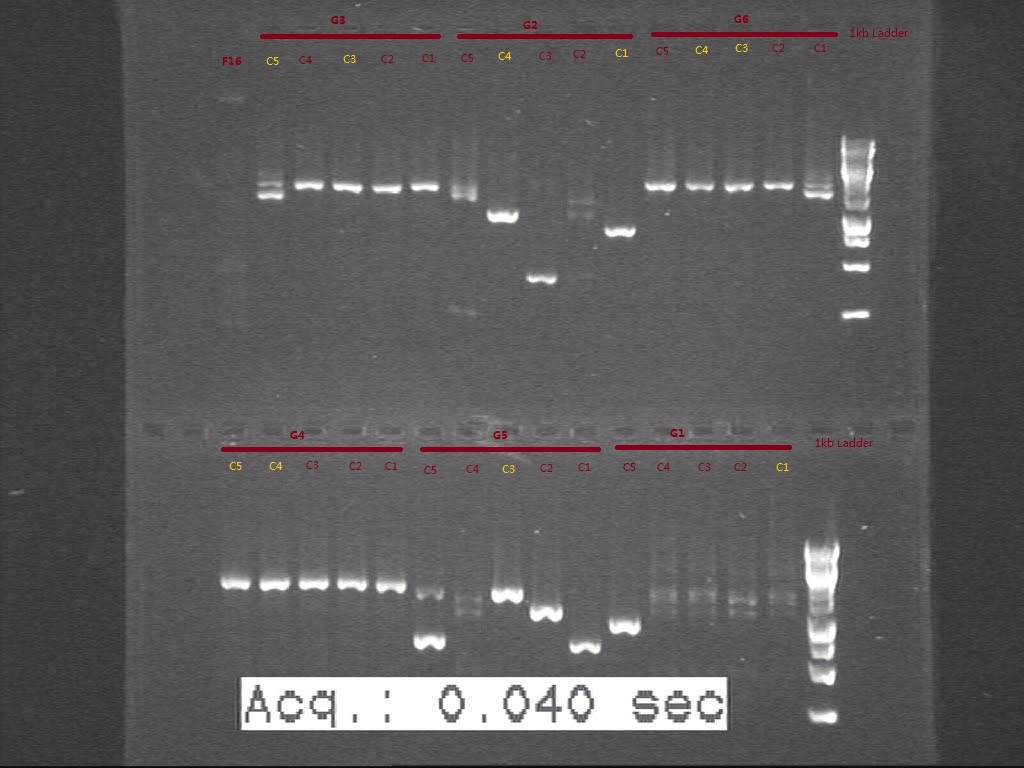
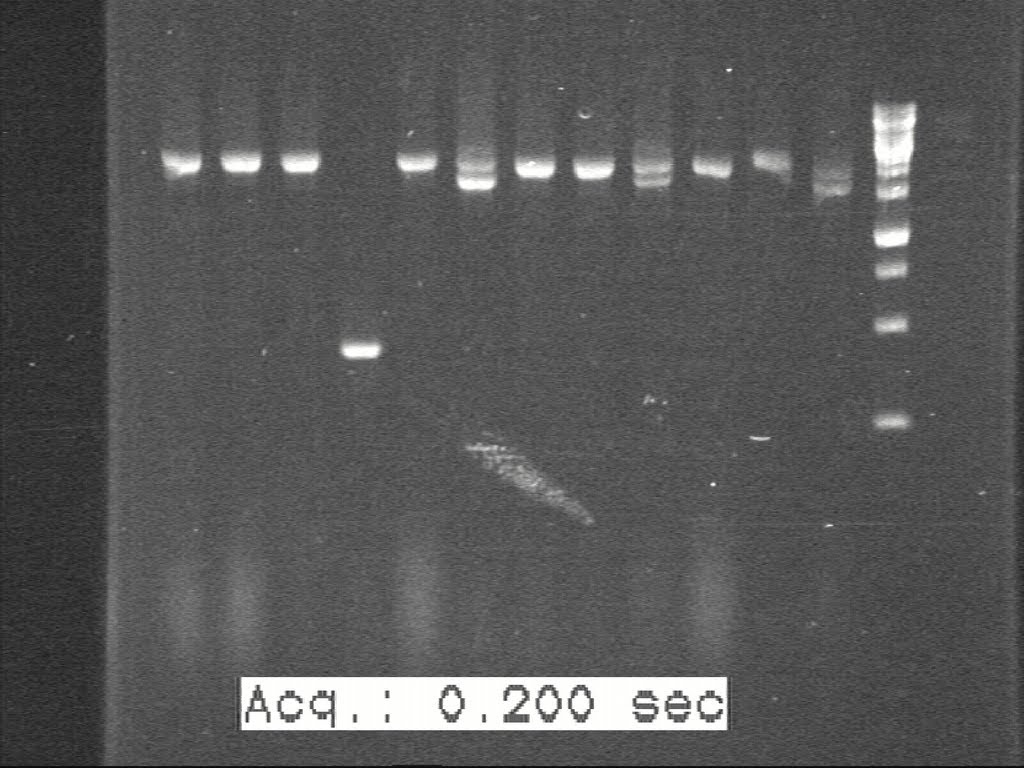
| + | 5 colonies were picked from each of those plates and sent to perform c-PCR. According to the band size on gel picture, 1 or 2 colonies of each set were selected to inoculate. | ||
| + | |||
| + | [[Image:Figure 3.jpg|500px|thumb|center|border|Figure 3]] | ||
| + | |||
| + | |||
| + | Again, inoculated samples were sent to perform plasmid extraction and measure the plasmid concentration. | ||
| + | |||
| + | Table . Plasmid concentration of G sets and F16. | ||
| + | |||
| + | Plasmids were digested with proper enzymes bought from Sigma-Aldrich and run for gel to confirm the size. | ||
| + | |||
| + | [[Image:Figure 4.jpg|500px|thumb|center|border|Figure 4 Gel picture shows the length of digested G sets]] | ||
| + | |||
| + | |||
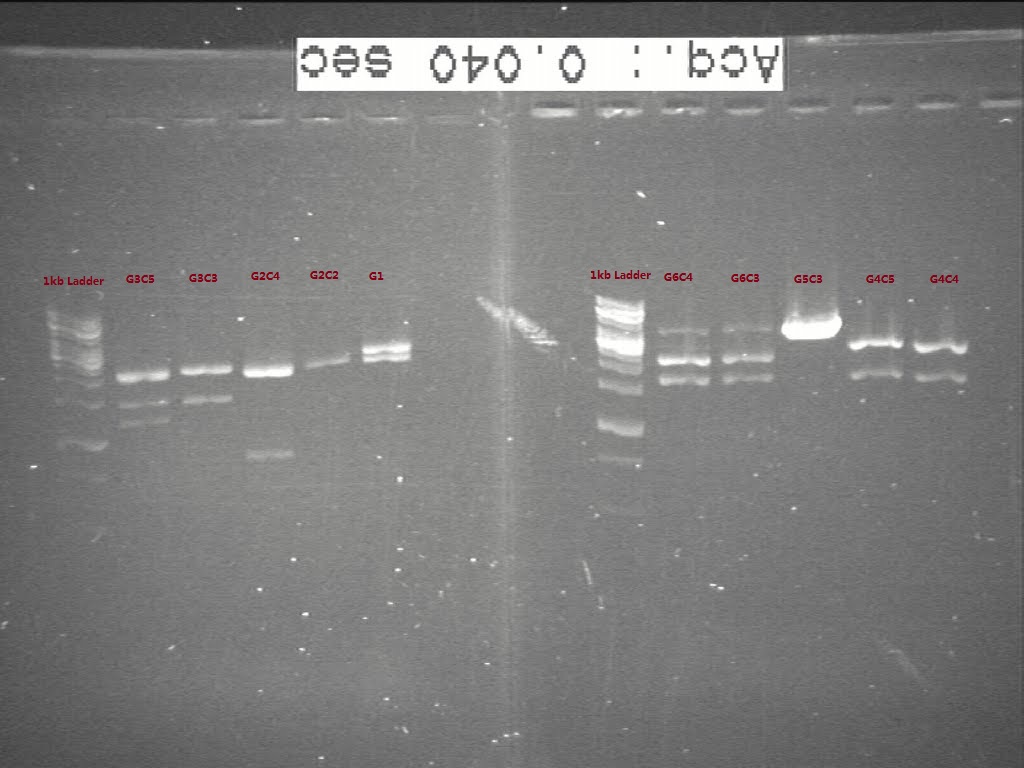
| + | From the gel, we think G1 and G2C4 seems having the right biobricks in size. G5 seems not digested well, though the plasmid seems in right size. The colonies we picked for G3, G4, G6 seems in wrong size according to c-PCR. Another transformation is necessary for G3, G4, and G6 and the starting biobricks need to be checked again by gel. | ||
| + | |||
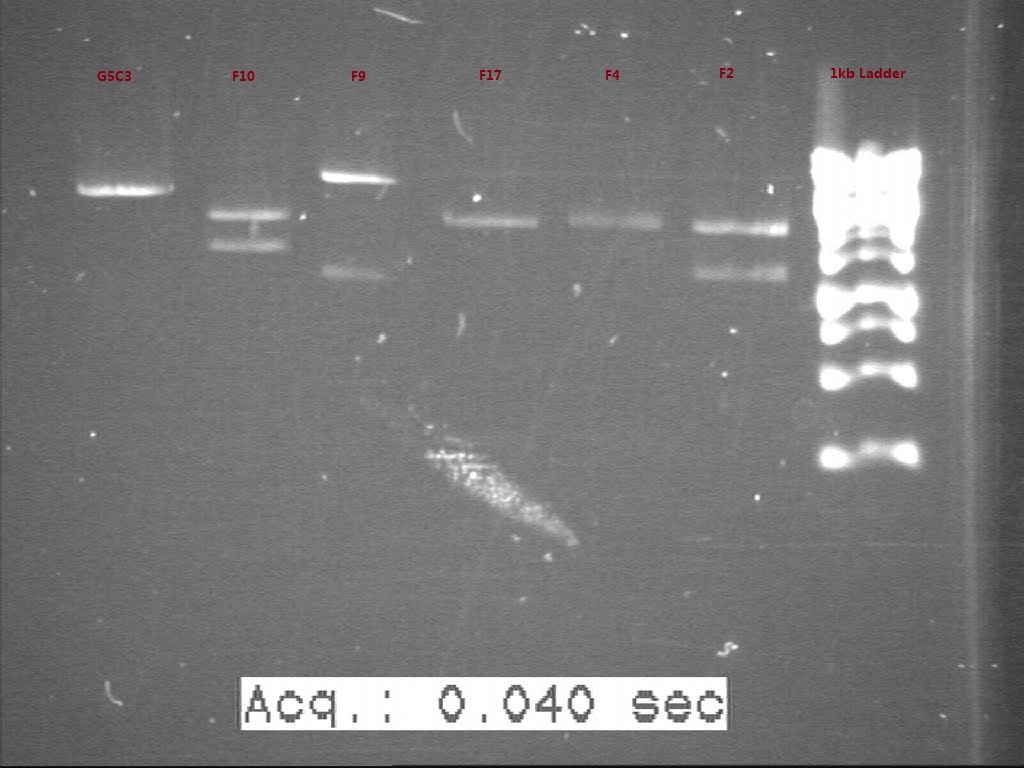
| + | [[Image:Figure 5.jpg|500px|thumb|center|border|Figure 5 Gel result to verify length of G5C3 and the starting material of G3, G4, and G6 ]] | ||
| + | |||
| + | |||
| + | All the starting material of G3, G4, and G6 seems good, though G5 C3 doesn’t seems in the right length. So a second transformation was done for G3, G4, G5, G6. After transformation, the cells were plated on the proper antibiotic plates. | ||
| + | 12 colonies were picked from each plates and sent to perform c-PCR. The c-PCR result was shown on the gel picture. | ||
| + | |||
| + | [[Image:Figure 6.jpg|500px|thumb|center|border|Figure 6 Gel picture shows the c-PCR result of second time transformation product of G3. The samples marked in orange were thought in right size and proceed to inoculate ]] | ||
| + | |||
| + | [[Image:Figure 7.jpg|500px|thumb|center|border|Figure 7 Gel picture shows the c-PCR result of second time transformation product of G4. The samples marked in orange were thought in right size and proceed to inoculate.]] | ||
| + | |||
| + | |||
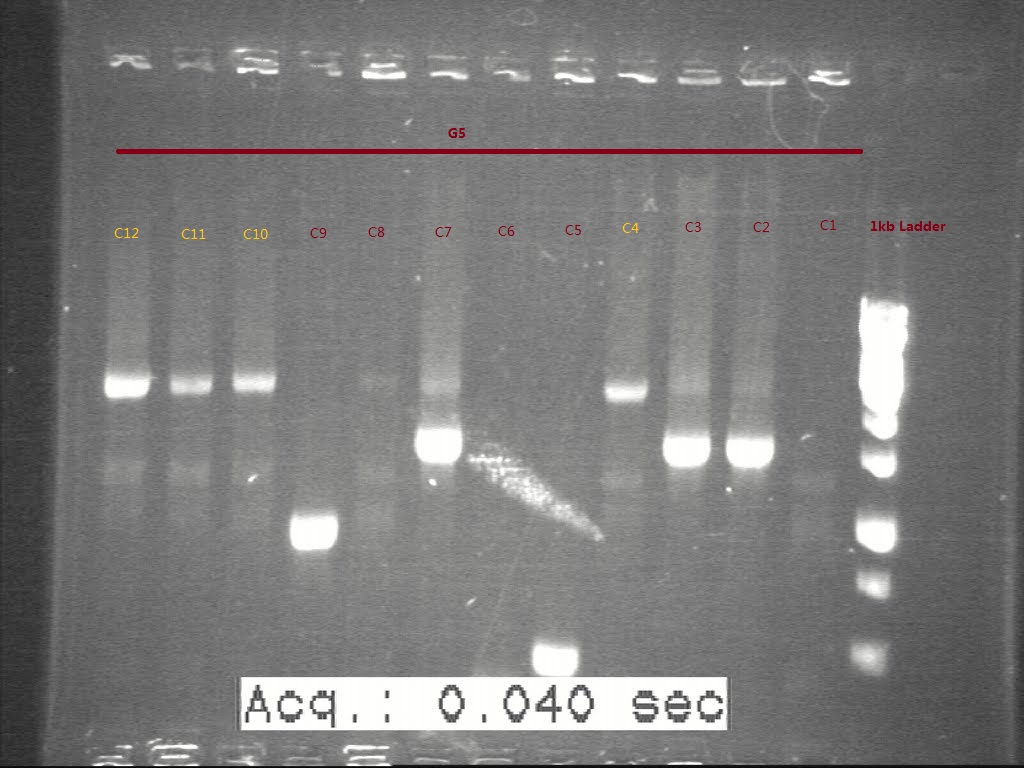
| + | [[Image:Figure 8.jpg|500px|thumb|center|border|Figure 8 Gel picture shows the c-PCR result of second time transformation product of G5. The samples marked in orange were thought in right size and proceed to inoculate.]] | ||
| + | |||
| + | [[Image:Figure 9.jpg|500px|thumb|center|border|Figure 9 Gel picture shows the c-PCR result of second time transformation product of G6. The samples marked in orange were thought in right size and proceed to inoculate.]] | ||
| + | |||
| + | |||
| + | Selected colonies were inoculated and performed plasmid extraction on the second day. The plasmid concentration was measured and result is showed in Table 4 | ||
| + | |||
| + | Table . Plasmid concentration of G3, G4, G6 sets | ||
| + | |||
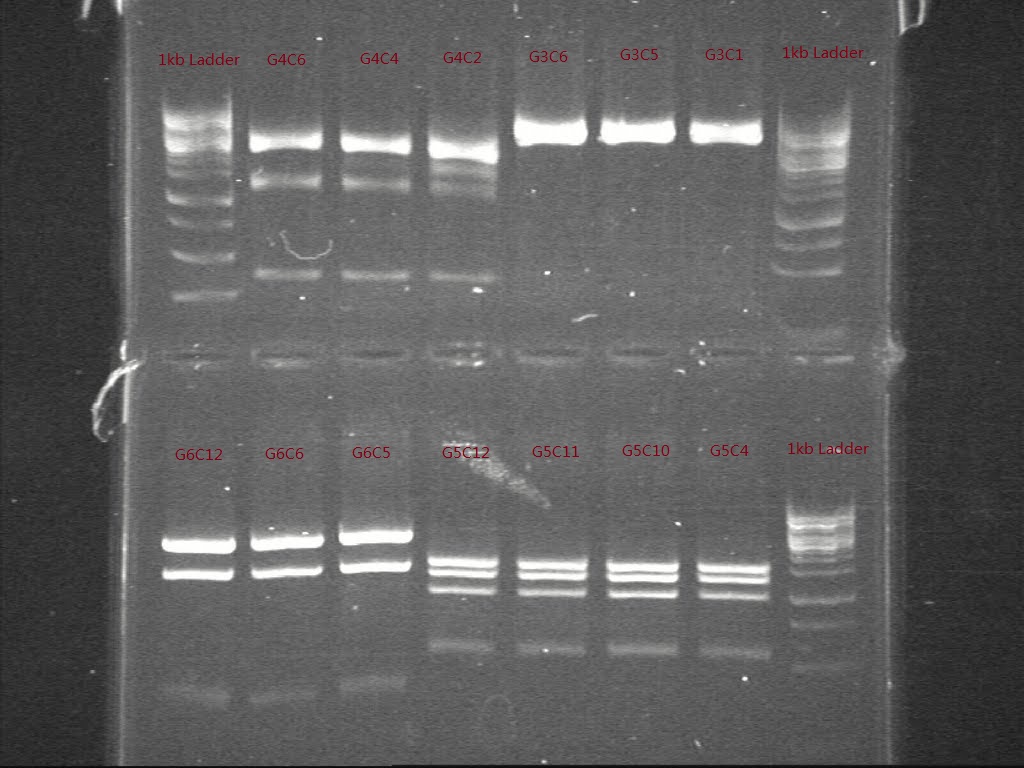
| + | For further confirm the construction of G sets, we decided to use some specific digestion and run the gel to verify the special sites on each construct. The digestion enzymes were chosen by using Gentle. | ||
| + | |||
| + | Table . Specific enzymes chosen for each construct and the expected length of all digested parts | ||
| + | |||
| + | [[Image:Figure 10.jpg|500px|thumb|center|border|Figure 10 Gel picture shows the result of specific digestion of G3, G4, G5 and G6.]] | ||
| + | |||
| + | |||
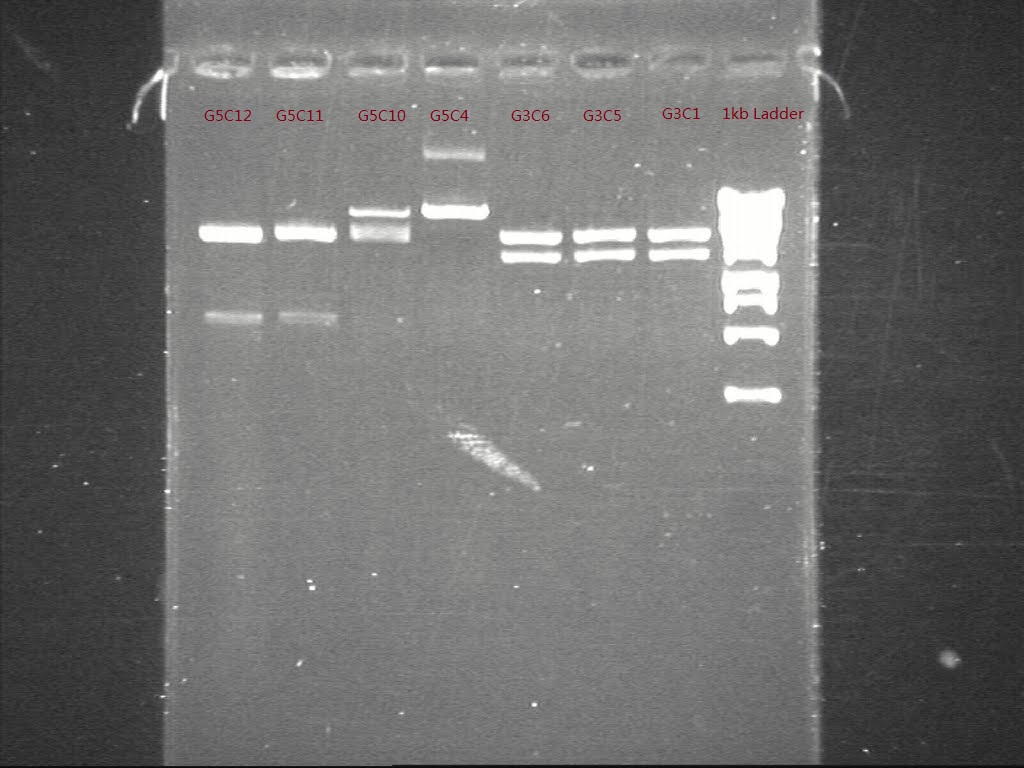
| + | Only samples from G4 and G6 showed in the expected length and consistence. A second specific digestion were performed to samples of G3 and G5 | ||
| + | |||
| + | [[Image:Figure 11.jpg|500px|thumb|center|border|Figure 11 Gel picture shows the result of specifc digestion of G3 and G5]] | ||
| + | |||
| + | Comparing the expected length of digested parts, G3C5, G4C6, G5C12 and G6C6 were digested and ligated according to the plan | ||
Revision as of 14:00, 24 October 2010



Week-10
Construction of G1- G7
5 colonies were picked from each of those plates and sent to perform c-PCR. According to the band size on gel picture, 1 or 2 colonies of each set were selected to inoculate.
Again, inoculated samples were sent to perform plasmid extraction and measure the plasmid concentration.
Table . Plasmid concentration of G sets and F16.
Plasmids were digested with proper enzymes bought from Sigma-Aldrich and run for gel to confirm the size.
From the gel, we think G1 and G2C4 seems having the right biobricks in size. G5 seems not digested well, though the plasmid seems in right size. The colonies we picked for G3, G4, G6 seems in wrong size according to c-PCR. Another transformation is necessary for G3, G4, and G6 and the starting biobricks need to be checked again by gel.
All the starting material of G3, G4, and G6 seems good, though G5 C3 doesn’t seems in the right length. So a second transformation was done for G3, G4, G5, G6. After transformation, the cells were plated on the proper antibiotic plates.
12 colonies were picked from each plates and sent to perform c-PCR. The c-PCR result was shown on the gel picture.
Selected colonies were inoculated and performed plasmid extraction on the second day. The plasmid concentration was measured and result is showed in Table 4
Table . Plasmid concentration of G3, G4, G6 sets
For further confirm the construction of G sets, we decided to use some specific digestion and run the gel to verify the special sites on each construct. The digestion enzymes were chosen by using Gentle.
Table . Specific enzymes chosen for each construct and the expected length of all digested parts
Only samples from G4 and G6 showed in the expected length and consistence. A second specific digestion were performed to samples of G3 and G5
Comparing the expected length of digested parts, G3C5, G4C6, G5C12 and G6C6 were digested and ligated according to the plan
 "
"