Team:Calgary/6 September 2010
From 2010.igem.org
Emily Hicks (Talk | contribs) |
|||
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{CalgaryNotebookTemplate| | {{CalgaryNotebookTemplate| | ||
| - | + | Monday September 6, 2010| | |
| + | |||
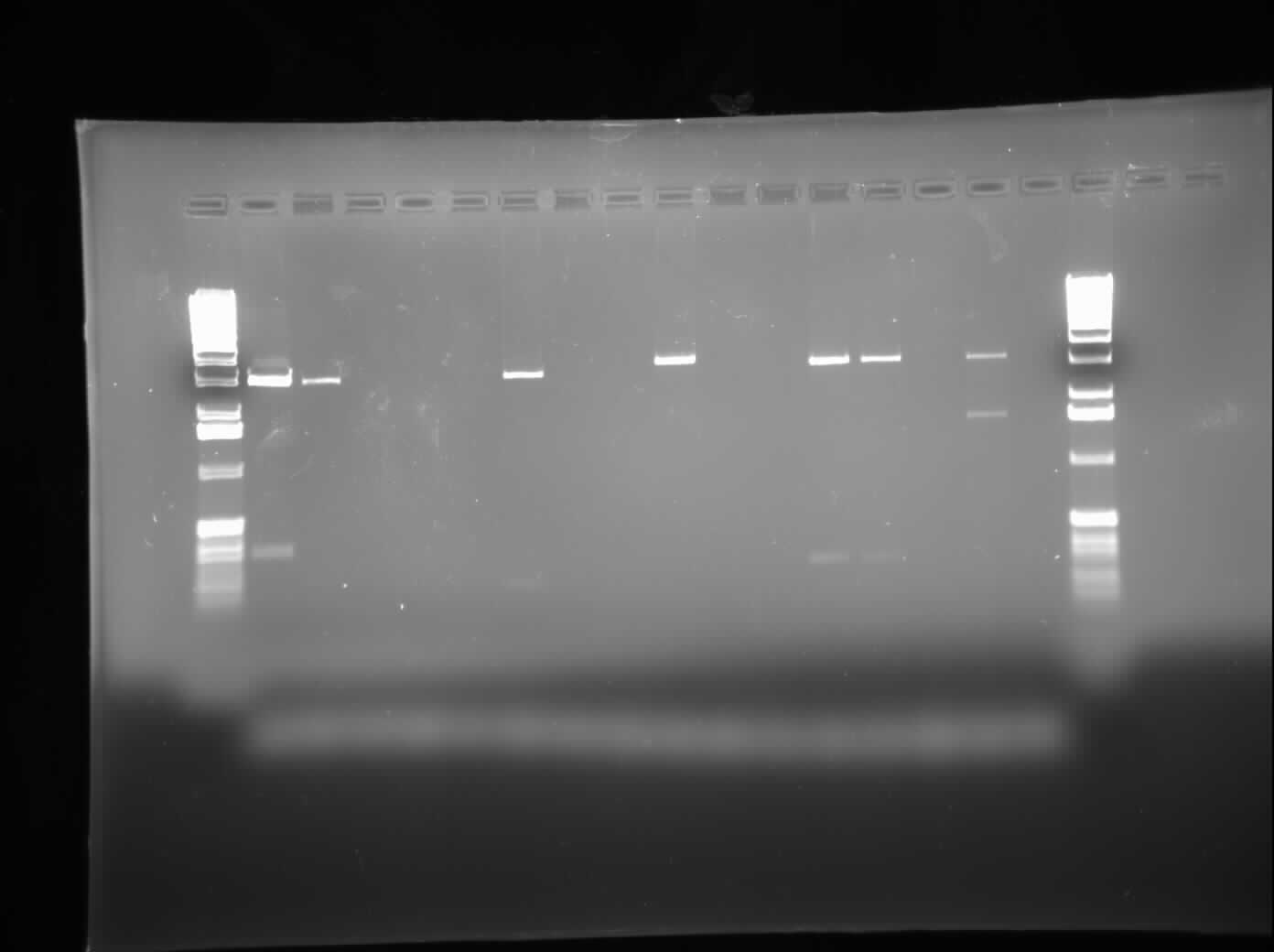
| + | [[Image:09.06.2010 CpxP & ibpAB New RD.jpg|thumb|400px|Chris's gel of the restriction digest of CpxP and ibpAB]] | ||
<u>Emily</u> | <u>Emily</u> | ||
| - | Today I ran PCR reactions of malE, malE31, malESSdel and malE31SSdel using the appropriate primers and an mgcl2 concentration of 2.5 as well as an annealing temperature of 55.5-57 degrees C. I got amplification of the expected bands (~1.2 KB) for the malE and malE31, however I once again got no bands for any of the SSdel stuff. These reactions will have to be further optimized tomorrow. Today a lot of work was also put into the presentation for aGEM this weekend in Edmonton. | + | Today I ran PCR reactions of malE, malE31, malESSdel and malE31SSdel using the appropriate primers and an mgcl2 concentration of 2.5 as well as an annealing temperature of 55.5-57 degrees C. I got amplification of the expected bands (~1.2 KB) for the malE and malE31, however I once again got no bands for any of the SSdel stuff. These reactions will have to be further optimized tomorrow. Today I also induced my I0500-I13504 cultures as well as Himika's cpxR reporter with malE31 circuit construct cultures with varying concentrations of Arabinose. We will be looking at GFP and RFP output for these tomorrow. Today a lot of work was also put into the presentation for aGEM this weekend in Edmonton. |
| + | |||
| + | |||
| + | <u>Jeremy</u> | ||
| + | |||
| + | Today I ran a gel with biobricked pRFP. From there I tried a gel extraction following the procedure in the protocols section. Unfortunately it didn't work, the spectophotometer revealed almost no DNA. I also did overnights for MalE, MalE31, nlpE in the TR49 TR50 Ravio cells. The majority of the day was spent working on the presentation animation and the first slide. | ||
| + | |||
| + | |||
| + | <u>Himika</u> | ||
| + | |||
| + | Today I did a PCR of I0500-B0034-MalE31 with different permutations of Dave's stuff and iGEM stuff to find out if something was contaminated. I also worked on the presentation for aGEM. | ||
| + | |||
| + | |||
| + | <u>Chris</u> | ||
| + | |||
| + | Today, I ran a 1.0% agarose gel electrophoresis of the restriction digests conducted yesterday on various ibpAB and CpxP parts. As well, I did plasmid preparations using the Sigma Aldrich Plasmid Preparation kit on the plasmids containing MalE and MalE31 with signal sequence deleted. Finally, I set up a regular PCR to try and verify the contents of the restriction digested parts. The gel was loaded with CpxP in Lanes 1-6, 8, 10, and 13; ibpAB in Lanes 7, 9, 11, and 12; and a negative control in Lane 14. The results can be seen on the right. | ||
}} | }} | ||
Latest revision as of 03:45, 17 October 2010

Monday September 6, 2010
Emily
Today I ran PCR reactions of malE, malE31, malESSdel and malE31SSdel using the appropriate primers and an mgcl2 concentration of 2.5 as well as an annealing temperature of 55.5-57 degrees C. I got amplification of the expected bands (~1.2 KB) for the malE and malE31, however I once again got no bands for any of the SSdel stuff. These reactions will have to be further optimized tomorrow. Today I also induced my I0500-I13504 cultures as well as Himika's cpxR reporter with malE31 circuit construct cultures with varying concentrations of Arabinose. We will be looking at GFP and RFP output for these tomorrow. Today a lot of work was also put into the presentation for aGEM this weekend in Edmonton.
Jeremy
Today I ran a gel with biobricked pRFP. From there I tried a gel extraction following the procedure in the protocols section. Unfortunately it didn't work, the spectophotometer revealed almost no DNA. I also did overnights for MalE, MalE31, nlpE in the TR49 TR50 Ravio cells. The majority of the day was spent working on the presentation animation and the first slide.
Himika
Today I did a PCR of I0500-B0034-MalE31 with different permutations of Dave's stuff and iGEM stuff to find out if something was contaminated. I also worked on the presentation for aGEM.
Chris
Today, I ran a 1.0% agarose gel electrophoresis of the restriction digests conducted yesterday on various ibpAB and CpxP parts. As well, I did plasmid preparations using the Sigma Aldrich Plasmid Preparation kit on the plasmids containing MalE and MalE31 with signal sequence deleted. Finally, I set up a regular PCR to try and verify the contents of the restriction digested parts. The gel was loaded with CpxP in Lanes 1-6, 8, 10, and 13; ibpAB in Lanes 7, 9, 11, and 12; and a negative control in Lane 14. The results can be seen on the right.
 "
"