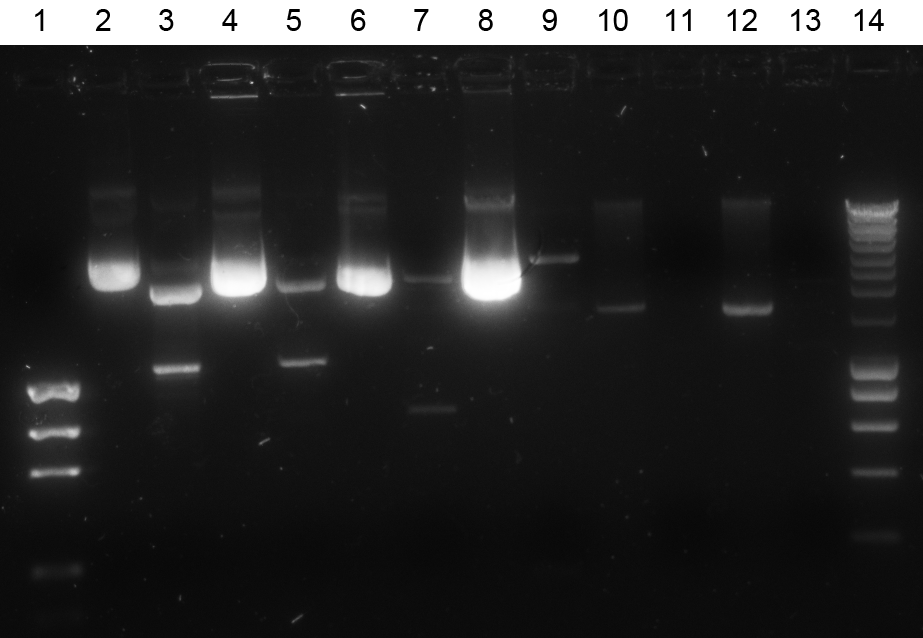Team:TU Delft/Lablog
From 2010.igem.org
All blog posts about lab work
Suits
The first custom made suits arrived from our sponsor SuitSupply. We're going to look awesome during the Jamboree.
Lab work
In the lab things are also getting started: Thias, Hugo and Pieter worked on preparation of the Top10 competent cells. These will be our workhorses for the next couple of weeks.
DNA order
After the relaxing jam session of yesterday, we all head back to the lab/office. Besides planning the trip to Paris workshop for this weekend, we also spend some time on setting up the first transformations. Also new possible sponsors are addressed. We are still on the hunt!
The ordering invoice for the DNA synthesis are signed and on their way to Mr Gene. Of course we added their logo to our webpages [http://www.igemtudelft.nl igemtudelft.nl] and [http://tiny.cc/igemtudelft Facebook] and [http://twitter.com/igemtudelft2010 Twitter]. And all team members have become fans on the network sites.
Lab work
Antibiotica stocks
Hugo, Ramon and Pieter made the antibiotic stock solutions. Though it is kinda hard to solubilize Chloramphenicol in water. Fortunately we have Hugo, the vortexman, on our team!
BioBrick stocks
We suspended some BioBricks from the distribution plate that we are planning to use in our project. Some of the BioBricks contain RFP. The red colour in our epps really looked nice! De BioBricks that we suspended were:
- [http://partsregistry.org/Part:pSB1A3 pSB1A3]
- [http://partsregistry.org/Part:pSB1K3 pSB1K3]
- [http://partsregistry.org/Part:pSB1T3 pSB1T3]
- [http://partsregistry.org/Part:pSB1C3 pSB1C3]
- [http://partsregistry.org/Part:pSB3T5 pSB3T5]
- [http://partsregistry.org/Part:pSB3C5 pSB3C5]
- [http://partsregistry.org/Part:I13401 I13401]
- [http://partsregistry.org/Part:B0015 B0015]
- [http://partsregistry.org/Part:B0032 B0032]
- [http://partsregistry.org/Part:B0034 B0034]
- [http://partsregistry.org/Part:E0040 E0040]
- [http://partsregistry.org/Part:E0240 E0240]
- [http://partsregistry.org/Part:E0422 E0422]
- [http://partsregistry.org/Part:J61100 J61100]
- [http://partsregistry.org/Part:J61101 J61101]
- [http://partsregistry.org/Part:J61107 J61107]
- [http://partsregistry.org/Part:J61117 J61117]
- [http://partsregistry.org/Part:J61127 J61127]
- [http://partsregistry.org/Part:J23109 J23109]
- [http://partsregistry.org/Part:R0010 R0010]
- [http://partsregistry.org/Part:R0011 R0011]
The concentration of the suspended BioBricks is not sufficient for all our plans. To make more plasmid, we are going to transform the BioBricks, grow colonies and isolate the plasmid.
Thias is also preparing for PCR amplification of a few biobricks. Esengül explained the mysteries of the magnificant PCR machines.
Quotes of the day
(While punching the first hole in the first biobrick plate ever)
Thias: What's in that bottle?
(something goes wrong)
Hugo: SHIT!
Eppy
Pieter just got a little present from Mariska van Ham :-) He's so happy with is eppy!
Today Nadine & Luke are learning how to work with HTML.
Lab work
BioBrick stocks
A number of Biobricks from the DNA distribution plates were suspended yesterday. Hugo and Kira did a transformation with 2 μL of the following BioBricks (antibiotic):
- pSB3T5 (Tetracycline)
- pSB1T3 (Tetracycline)
- pSB1C3 (Chloramphenicol)
- pSB3C5 (Chloramphenicol)
- pSB1K3 (Kanamycin)
- pSB1A3 (Ampicillin)
- I13401 (Ampicillin)
Because our own competent cells were not good enough we used commercially available competent Top10 cells (Invitrogen). The cells were plated on LB-agar with the appropriate antibiotic. Biobricks plated We will have to wait until tomorrow to see if they were successful or not. Hopefully we will see some colonies tomorrow. Wish us luck!
Thias PCRed some BioBricks with various primer concentrations and at different temperatures to see which PCR method would work best. The PCR products were tested on a gel, and we saw a difference between the primer concentrations and the temperatures used.
Media and Solutions
By Hugo
Nadine and I are planing to revive our Ps. putida strain (muahaha). So, we prepared some solid media for that purpose: LB agar and E2 agar.
- Tryptone 10g
- Yeast extract 5 g
- NaCl 10 g
- Agar 15 g
- Water, as much as you need for 1 L of culture medium
- (NH4)2HPO4 10 g
- K2HPO4 5 g
- Na2SO4 0.5g
- 1 ml of MT stock solution
- 1 ml of 1M MgSO4
- Water, as much as you need for 1 L of culture medium
If you want to prepare the solid medium, add 15 g of agar per liter of medium.
MT stock solution
- FeSO4 7H2O 2.8 g
- MnCl2 0.19g
- CoCl2 6H2O 2.8 g
- CaCl2 4H2O 1.84g
- ZnSO4 7H2O 0.28 g
- CuSO4 0.16g
WARNING: Dear iGEM people be aware that the original E2 composition is the following (Lageveen et al., 1988):
- KH2PO4 3.7 g
- K2HPO4 7.5 g
- NaNH4HPO4.4H2O 3.5 g
- Citrate 5 g / 12 g
- 1 M MgSO4 . 7H2O 1 ml
- MT stock solution 1 ml
- FeSO4.7H2O 2.8 g
- MnSO4.H2O 2 g
- CoCl2.6H2O 2.8 g
- CaCl2.2H2O 1.48 g
- ZnSO4.7H2O 0.28 g
- CuCl2.2H2O 0.16 g
Why did we change the medium?
First, we lack of the weird salt NaNH4HPO4.4H2O, we want to save some money by making some engineering in the culture medium. Instead of the main salts for E2 we are using the salts recommended by [http://www.lgcstandards-atcc.org/ American Type Culture Collection] (ATCC) in the optimal medium for Ps. putida, this mineral medium is also known as P1. Find the original medium composition [http://www.lgcstandards-atcc.org/Attachments/4091.pdf here].
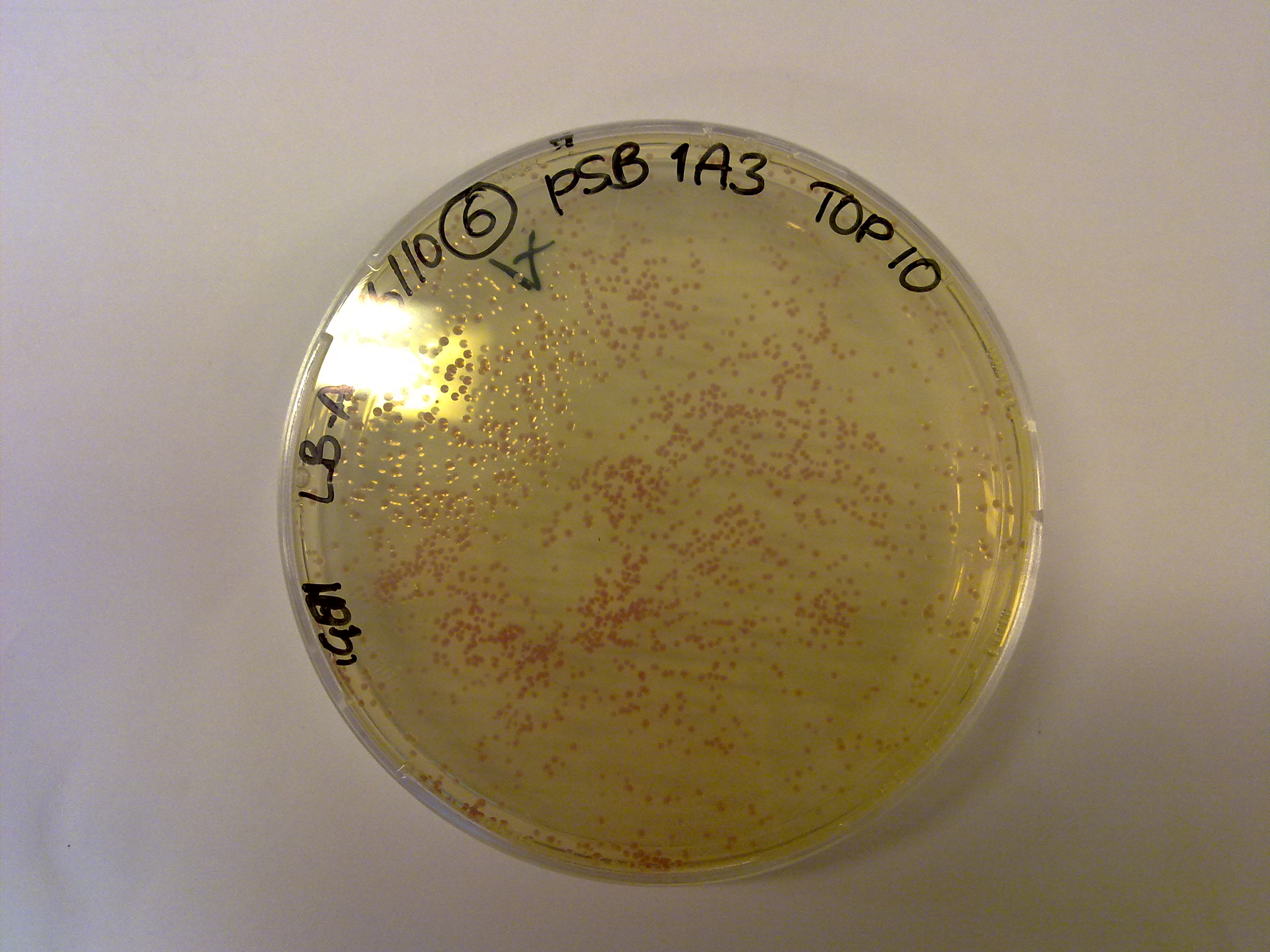
Lab work
BioBrick stocks
We got the first colonies, and they're red! Hurray!
Yesterday we grew colonies containing the vector backbones (which contain RFP). Today one colony from each plate was picked and cultivated in liquid (4 mL LB + 4 μL antibiotic) as well as solid LB medium (+20 μL antibiotic) with the corresponding antibiotic to grow overnight.
Thias purified the PCR products.
Strain Characterization
By Hugo
Today we tried to revive our strain. We added some water to the lyophilized powder. And we plated some microliters (100 ul) from the dispersion obtained on LB agar, E2 agar with octane and E2 with glucose. Let's hope for the best!. Growing temperature: 30º C
Lab work
BioBrick stocks
Today we performed a Qiagen Mini-prep plasmid isolation of the vectors we grew overnight. We saved 3 mL of the bacterial cells to make -80 °C stocks.
The following plasmid concentrations were obtained:
| BioBrick | Concentration (ng/μL) |
| pSB3T5 | 266.0 |
| pSB1T3 | 414.0 |
| pSB1C3 | 284.7 |
| pSB3C5 | 329.3 |
| pSB1K3 | 270.1 |
| pSB1A3 | 271.9 |
| I13401 | 232.1 |
Characterization of Anderson RBS sequences
Next to that we also started digesting the first parts we will be characterizing.
Strain Characterization
By Hugo
I prepared the cell bank according to the following procedure:
- Spin 5 ml of the overnight culture
- Add 500 ul of LB medium
- Add 500 ul of 80% glycerol
- Store the tube at -80ºC
Now, we have a cell bank ready to be used!!!
Spring Workshop
Almost the whole team left for the iGEM spring workshop in Évry, Paris. The drive for some of us was a bit laborious (traffic jams at the middle of the night/closed roads etc.), and others actually watched the World Cup soccer match... But in the end we all arrived at the hotel in Évry little before 2 AM, Saturday morning, and got some well deserved sleep.
Cleaning
After all the media attention, we had a nice slow day. We finished some small experiments and reorganized our iGEM room. We hide all the stuff from last years team in a closet so now our room is nice and tidy.
We also booked a hotel in Boston!
Lab work
Emulsifier
Pieter worked on the emulsifying activity assay. Four samples were tested:
1. Mix, 0.02 mL of substrate, 0.1- 0.5 mL of sample.
| # | Amount of SDS (%) | Hexalkane (mL) |
| 1 | 0 | 0 |
| 2 | 0 | 0.02 |
| 3 | 0.01 | 0.02 |
| 4 | 1 | 0.02 |
2. Tris buffer (pH 8) was added to a total volume of 7.5 ml.
3. Shake 1 hour with 200 strokes/min at 30°C
4. Measure absorbance OD600
| # | A600 |
| 1 | 0 |
| 2 | 0 |
| 3 | 0.4 |
| 4 | 0.2 |
Biobricks
By Hugo
I've got some colonies in the plates, however there's contamination with some yellow colonies :s Probably, we will have to prepare a new stock of competent cells.
Lab work
Characterization of Anderson RBS sequences
Restriction digestion of plasmid backbone pSB1T3.
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA | Needed fragment |
| 1 | 1.0 μg pSB1T3 | EcoRI | PstI | H (Roche) | ✗ | ‘E–linear pSB1T3–P’ |
In stead of 10 U/μL restriction enzyme we used 1 U/μL. The digested pSB1T3 was cut from gel and isolate the band. However, the gel extraction kit didn’t work as well as we had hoped, it turned out to be the DNA disappearing kit. Tomorrow a new day.
Lab work
Characterization of Anderson RBS sequences
We tried to compensate for yesterdays delays. Thias performed a PCR reaction on short BioBrick inserts of J61100, J61101, J61107, J61117, J61127 and B0034. We wanted to investigate if this would save time in the long-run compared to transforming and plasmid isolation. The PCR reaction was run at varying annealing temperatures to figure out which temperature gave the best result.
After analysis on gel we deduced that a temperature of 50 °C was optimal.
At the end of the day we performed a Restriction digestion reaction on the amplified PCR products as well as the previously isolated I13401 and pSB3C5:
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA | Needed fragment |
| 1 | 1.0 μg pSB1T3 | EcoRI | PstI | H (Roche) | ✗ | ‘X–linear pSB1T3–P’ |
In stead of 10 U/μL restriction enzyme we used 1 U/μL. Mixtures were incubated at 37 °C for 2.5 hours and stored at 4 °C overnight.
Lab work
Characterization of Anderson RBS sequences
Ligations were performed using the overnight digested BioBricks. The following ligation reactions was performed:
| # | BioBrick | Fragment 1 | Fragment 2 | Recipient vector |
| 1 | 4 μL μL ‘E–J1100–S’ | 3 μL μL ‘X–I10341–P’ | 1.5 μL ‘E–linear pSB1T3–P’ | |
| 2 | 4 μL μL ‘E–J1101–S’ | 3 μL μL ‘X–I10341–P’ | 1.5 μL ‘E–linear pSB1T3–P’ | |
| 3 | 4 μL μL ‘E–J1107–S’ | 3 μL μL ‘X–I10341–P’ | 1.5 μL ‘E–linear pSB1T3–P’ | |
| 4 | 4 μL μL ‘E–J1117–S’ | 3 μL μL ‘X–I10341–P’ | 1.5 μL ‘E–linear pSB1T3–P’ | |
| 5 | 4 μL μL ‘E–J1127–S’ | 3 μL μL ‘X–I10341–P’ | 1.5 μL ‘E-linear pSB1T3–P’ | |
| 6 | negative control | - | 3 μL μL ‘X–I10341–P’ | 1.5 μL ‘E–linear pSB1T3–P’ |
We adhered to the 3:1 insert:plasmid ratio when determining the volumes of DNA added. The mixtures were incubated overnight at 16 °C.
Media and solutions
By Hugo
The recipe is as follows:
| Compound | Amount required | Final concentration |
| Bromophenol blue | 0.025 g | 0.25% w/v |
| Xylene Cyanol FF | 0.025 g | 0.25% w/v |
| Ficoll (type 400: Pharmacia) | 1.5 g | 15% w/v |
| Water | As much as you need for 10 ml |
WARNING: THE POWDERS ARE ELECTROSTATIC. WEAR GLOVES AND CLEAN ALL THE PLACE WITH ALCOHOL (70% v/v)... SERIOUSLY, THE BLUE STUFF IS EVERYWHERE!!!
Lab work
BioBrick stocks
To make more plasmid, we transformed 1 μL of 14 BioBricks from the distribution plates:
- B0015
- B0032
- B0034
- E0040
- E0240
- E0422
- J61100
- J61101
- J61107
- J61117
- J61127
- J23109
- R0010
- R0011
Chess
We have several chess players in our teams. Meanwhile a hard competition is going on in our iGEM room.
Quote of the Day
"Sometimes turtles can turn into ninjas!" -Luke and Ramon playing chess way too defensive
World Championship
We finished our experiments before 16.00 h, so we could watch the game in our iGEM office. Go Orange! Stop sound
Lab work
We are a bit short handed in the lab this week, because most team members have exams. Considering the fine weather outside, it is not bad to have a slow week.
BioBrick stocks
All the transformations of yesterday containing BioBricks form the distribution plate gave colonies. We pick a colony of every plate and grow them overnight in 50 mL LB medium containing Ampicillin.
Pseudomonas Putida GPO1
Today we cultivated Pseudomonas Putida GPO1 on different substrates to test on what carbon source it can grow:
1. E2 medium without carbon source
2. E2 medium + 1% LB
3. E2 medium + 10% LB
4. E2 medium + 1% octane
5. E2 medium + 10% octane
Lab work
BioBrick stocks
We harvested the 50 mL bacterial cells of the 14 BioBricks. We used 3 mL of the baceterial cells to make -80 °C stocks. With the rest we performed a Qiagen Midi-prep plasmid isolation.
The following plasmid concentrations were obtained:
| BioBrick | Concentration (ng/μL) |
| B0015 | 360.5 |
| B0032 | 80.4 |
| B0034 | 181.7 |
| E0040 | 66.0 |
| E0240 | 305.6 |
| E0422 | 411.0 |
| J61100 | 128.7 |
| J61101 | 167.7 |
| J61107 | 195.9 |
| J61117 | 62.7 |
| J61127 | 33.8 |
| R0010 | 40.0 |
| R0011 | 42.0 |
Pseudomonas Putida GPO1
Pseudomonas Putida GPO1 is growing 1 day on different substrates. We measured absorbance of the Pseudomonas Putida to see on which conditions the strain survives. Absorbance measurements
Lab Work
Pseudomonas Putida GPO1
The second day of the grow process of Pseudomonas Putida GPO1
| Sample | Medium | day 1 = 30 June | day 2 = 1 July |
| no cells | E2 without carbon | 0.00 | 0.00 |
| no cells | LB | 0.00 | 0.00 |
| no cells | E2 + 1% LB | 0.00 | 0.01 |
| Pseudomonas Putida GPO1 * | E2 + 1% LB | 0.01 | 0.05 |
| no cells | E2 + 10% LB | 0.00 | 0.00 |
| Pseudomonas Putida GPO1 * | E2 + 10% LB | 0.34 | 0.19 |
| no cells | E2 + 1% octane | 0.00 | 0.00 |
| Pseudomonas Putida GPO1 * | E2 + 1% octane | 0.00 | 0.00 |
| Pseudomonas Putida GPO1 * | E2 + 10% octane | 0.29 | 0.18 |
* average of 3 colonies
Pseudomonas Putida GPO1 can grow on 10% octane as carbon source.
Competent cells
Top10 cells were cultivated for making competent cells.
Lab work
BioBrick stocks
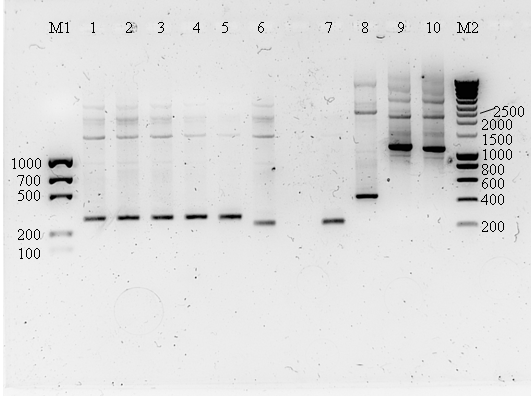
Today we performed Colony PCR with the transformants of the BioBricks to check if everything is what is says it is.
Results of the colony PCR on 1% agarose gel
Lane description:
| # | Description | Expected lenght (bp) | Primers | Status |
| M1 | EZ load percision | n/a | n/a | n/a |
| 1 | J61100 | 293 | G00100 + G00101 | ✓ |
| 2 | J61101 | 293 | G00100 + G00101 | ✓ |
| 3 | J61107 | 293 | G00100 + G00101 | ✓ |
| 4 | J61117 | 293 | G00100 + G00101 | ✓ |
| 5 | J61127 | 293 | G00100 + G00101 | ✓ |
| 6 | B0034 | 250 | G00100 + G00101 | ✓ |
| 7 | J23109 | G00100 + G00101 | ✓ | |
| 8 | B0015 | 445 | G00100 + G00101 | ✓ |
| 9 | E0422 | 1155 | G00100 + G00101 | ✓ |
| 10 | E0240 | 1114 | G00100 + G00101 | ✓ |
| M2 | Smartladder | n/a | n/a | n/a |
Competent cells
At the same time Eva is making more competent cells. We go through our competent cells so quickly. To test the efficiency of the competent cells, 100 ng standard pUC19 vector was transformed and plated on LB agar containing 100 μg/mL Ampicillin. The plates were incubated on room temperature.
World Championship
We watched the World Cup game (the Netherlands - Brazil) in our iGEM room. Care was taken for many tasty snacks. Cake, chips and drinks Mmmmm.
Lab work
BioBrick stocks
Today we performed Colony PCR with the transformants of the BioBricks to check if everything is what is says it is.
Results of the colony PCR on 1% agarose gel
Lane description:
| # | Description | Expected lenght (bp) | Primers | Status |
| M1 | EZ load percision | n/a | n/a | n/a |
| 1 | J61100 | 293 | G00100 + G00101 | ✓ |
| 2 | J61101 | 293 | G00100 + G00101 | ✓ |
| 3 | J61107 | 293 | G00100 + G00101 | ✓ |
| 4 | J61117 | 293 | G00100 + G00101 | ✓ |
| 5 | J61127 | 293 | G00100 + G00101 | ✓ |
| 6 | B0034 | 250 | G00100 + G00101 | ✓ |
| 7 | J23109 | G00100 + G00101 | ✓ | |
| 8 | B0015 | 445 | G00100 + G00101 | ✓ |
| 9 | E0422 | 1155 | G00100 + G00101 | ✓ |
| 10 | E0240 | 1114 | G00100 + G00101 | ✓ |
| M2 | Smartladder | n/a | n/a | n/a |
Competent cells
At the same time Eva is making more competent cells. We go through our competent cells so quickly. To test the efficiency of the competent cells, 100 ng standard pUC19 vector was transformed and plated on LB agar containing 100 μg/mL Ampicillin. The plates were incubated on room temperature.
World Championship
We watched the World Cup game (the Netherlands - Brazil) in our iGEM room. Care was taken for many tasty snacks. Cake, chips and drinks Mmmmm.
Lab work
Ordered DNA stocks
The DNA from Mr Gene has finally arrived, now we're ready to build some biobricks!
We were so excited; we immediately dissolved the DNA in water and performed a transformation with 100 ng of the DNA and plated on the LB agar containing the appropriate antibiotic.
- AlkB2 (Ampicillin)
- rubA3 (Ampicillin)
- rubA4 (Ampicillin)
- rubB (Kanamycin)
- ladA (Ampicillin)
- ADH (Ampicillin)
- ALDH (Kanamycin)
- bbc1 (Ampicillin)
- AlnA (Ampicillin)
- OprG (Ampicillin)
- PalkS1-2 (Ampicillin)
- PalkB (Ampicillin)
- P(CaiF) (Ampenicillin)
- AlkS (Kanamycin)
- PhPFD-α (Ampicillin)
- PhPFD-β (Ampicillin)
Characterization of Anderson RBS sequences
Yesterday's purified PCR product of I13401 and the Anderson RBS Biobricks (J61100, J61101, J61107, J61117 and J61127) were digested:
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA | Needed fragment |
| C1 | 1.15 μg J61100 | SpeI | PstI | 2 (BioLabs) | ✓ | ‘S–J61100 pSB1A2–P’ |
| C2 | 1.68 μg J61101 | SpeI | PstI | 2 (BioLabs) | ✓ | ‘S–J61101 pSB1A2–P’ |
| C3 | 1.30 μg J61107 | SpeI | PstI | 2 (BioLabs) | ✓ | ‘S–J61107 pSB1A2–P’ |
| C4 | 0.95 μg J61117 | SpeI | PstI | 2 (BioLabs) | ✓ | ‘S–J61117 pSB1A2–P’ |
| C5 | 0.51 μg J61127 | SpeI | PstI | 2 (BioLabs) | ✓ | ‘S–J61127 pSB1A2–P’ |
| D1 | 2.59 μg I13401 | XbaI | PstI | 2 (BioLabs) | ✓ | ‘X–I13401–P’ |
| D2 | 2.65 μg I13401 | XbaI | PstI | 2 (BioLabs) | ✓ | ‘X–I13401–P’ |
The digestion products were purified using Roche's PCR Purifiation Kit and loaded onto a 1% agarose gel for comparison with the non-digested BioBricks:
Lane description:
| # | Description | Expected Length (bp) | Status |
| M1 | SmartLadder marker (5 μL) | n/a | n/a |
| 1 | Undigested J61100 in pSB1A2 | 2134 | ✓ |
| 3 | Undigested J61101 in pSB1A2 | 2134 | ✓ |
| 4 | J61100 + SpeI + PstI | 18, 2116 | ✓ |
| 5 | J61101 + SpeI + PstI | 18, 2116 | ✓ |
| 6 | Undigested J61107 in pSB1A2 | 2134 | ✓ |
| 7 | J61107 + SpeI + PstI | 18, 2116 | ✓ |
| 8 | Undigested J61117 in pSB1A2 | 2134 | ✓ |
| 9 | J61117 + SpeI + PstI | 18, 2116 | ✓ |
| 10 | Undigested J61127 in pSB1A2 | 2134 | ✓ |
| 11 | J61127 + SpeI + PstI | 18, 2116 | ✓ |
| 12 | Undigested I13401 in pSB1A2 | 2936 | ✓ |
| 13 | I13401 + XbaI + PstI | 883, 2053 | ✓ |
| 14 | empty | empty | empty |
| 15 | BioRad EZ Load | n/a | n/a |
Followed by over night ligation:
| # | BioBrick | Recipient plasmid | Fragment | Final volume |
| 1 | K398500A | 130 μg ‘S–J61100 pSB1A2–P’ | 154 μg ‘X-I13401-P’ | 26 μL |
| 2 | K398501A | 206 μg ‘S–J61101 pSB1A2–P’ | 247 μg ‘X-I13401-P’ | 25 μL |
| 3 | K398502A | 196 μg ‘S–J61107 pSB1A2–P’ | 247 μg ‘X-I13401-P’ | 25 μL |
| 4 | K398503A | 201 μg ‘S–J61117 pSB1A2–P’ | 247 μg ‘X-I13401-P’ | 25 μL |
| 5 | K398504A | 202 μg ‘S–J61127 pSB1A2–P’ | 247 μg ‘X-I13401-P’ | 25.5 μL |
| 6 | negative control | 123 μg ‘S–J61117 pSB1A2–P’ | - | 15 μL |
To all samples one-tenth of the total volume ligase buffer was added.
Emulsifier
Pieter is testing different condition for the emulsifying Assay. To determine the emulsifying activity we set up a calibration curve with SDS. He prepared the following samples:
| # | Hexane (mL) | Tris Buffer pH 8 (mL) | 10% SDS (mL) |
| B | 1 | 1.1 | 0 |
| 100 | 1 | 1 | 0.1 |
| 50 | 1 | 1.05 | 0.05 |
| 25 | 1 | 1.075 | 0.025 |
| 10 | 1 | 1.09 | 0.01 |
| 5 | 1 | 1.095 | 0.005 |
Kampioenen!!!!
Today the Dutch team will play the semi-final of the World Cup! Hear us cheer! Stop sound
Lab work
Ordered DNA stocks
All the colonies of the transformations of yesterday grew. At the end of the day we picked colonies and grew them overnight in 200 mL LB medium containing the appropriate antibiotic.
Characterization of Anderson RBS sequences
Transformation of yesterday's ligation product into competent TOP10 cells and over night incubation.
Lab work
Ordered DNA stocks
We harvested the 200 mL bacterial cells of the 16 DNA parts. We used 3 mL of the bacterial cells to make -80 °C stocks. With the rest we centrifuged at 4,000 rmp for 15 minutes and stored the pellets in the -20 °C.
Characterization of Anderson RBS sequences
Tuesday's ligation products of BioBricks K398500-K398504 yielded successful transformants. Single colony PCRs were performed and loaded onto a gel. The bands on the 1% agarose gel indicated the presence of inserts with the proper lengths:
Lane descriptions:
| # | Description | Expected Length (bp) | Primers | Status | Remarks |
| 1 | transformant #1 of ligation mix K398500 | 1158 | G00100 + G00101 | ✓ | Unexpected band at 2700 |
| 2 | transformant #2 of ligation mix K398500 | 1158 | G00100 + G00101 | ✓ | |
| 3 | transformant #1 of ligation mix K398501 | 1158 | G00100 + G00101 | ✓ | |
| 4 | transformant #2 of ligation mix K398501 | 1158 | G00100 + G00101 | ✓ | |
| 5 | transformant #1 of ligation mix K398502 | 1158 | G00100 + G00101 | ✓ | |
| 6 | transformant #2 of ligation mix K398502 | 1158 | G00100 + G00101 | ✓ | |
| 7 | transformant #1 of ligation mix K398503 | 1158 | G00100 + G00101 | ✓ | |
| 8 | transformant #2 of ligation mix K398503 | 1158 | G00100 + G00101 | ✓ | probably run of the gel |
| 9 | transformant #1 of ligation mix K398504 | 1158 | G00100 + G00101 | ✓ | |
| 10 | transformant #2 of ligation mix K398504 | 1158 | G00100 + G00101 | ✓ | Sample not fully loaded on gel |
| 11 | transformant #1 of ligation control | G00100 + G00101 | probably run of the gel | ||
| 12 | transformant #2 of ligation control | G00100 + G00101 | |||
| 13 | transformant #1 of digestion control | G00100 + G00101 | probably run of the gel | ||
| 14 | transformant #2 of digestion control | G00100 + G00101 | |||
| M1 | SmartLadder Marker | n/a | n/a | n/a |
The colonies belonging to lanes 2, 4, 6, 8 and 10 were plated out on AMP plates to yield reincultures for subsequent characterization experiments.
Alkane degradation
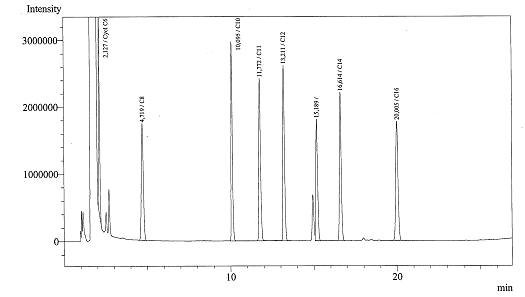
Today we got the Gas Chromatograph working, we could identify several peaks. Now we are ready for the real experiments!
Lab work
Ordered DNA stocks
Ramon and Eva are performing Qiagen Maxi-prep plasmid isolation of ordered DNA parts.
The following plasmid concentrations were obtained:
| DNA part | Concentration (ng/μL) |
| alkB2 | 147.3 |
| ruBA3 | 63.6 |
| rubA4 | 29.0 |
| rubR | 26.7 |
| ladA | 5011.2 |
| ADH | 178.8 |
| ALDH | 24.1 |
| bbc1 | 5.5 |
| AlnA | 357.4 |
| OprG | 197.3 |
| AlkS | 510.8 |
| PalkB | 61.5 |
| PalkS1-2 | 60.9 |
| P(Caif) | 9.7 |
| PhPFDα | 147.0 |
| PhPFDβ | 56.7 |
rubA4, rubR, ALDH, bbc1 and P(Caif) have a low concentration, so we going to repeat the plasmid isolation of these samples.
Lab work
Characterization of Anderson RBS sequences
E.coli Top10 strains containing the following composite BioBricks in pSB1A2 had been obtained:
| BioBrick | Composed of: |
| [http://partsregistry.org/Part:BBa_K398500 K398500] | J23100 + J61100 + I13401 |
| [http://partsregistry.org/Part:BBa_K398501 K398501] | J23100 + J61101 + I13401 |
| [http://partsregistry.org/Part:BBa_K398502 K398502] | J23100 + J61107 + I13401 |
| [http://partsregistry.org/Part:BBa_K398503 K398503] | J23100 + J61117 + I13401 |
| [http://partsregistry.org/Part:BBa_K398504 K398504] | J23100 + J61127 + I13401 |
To prepare for the fluorescence assay measurements the strains were grown in 3 mL LB medium containing 100 μg/mL AMP as well as in M9 minimal medium containing 0.4% glucose and 100 μg/mL AMP.
High copy number RBS characterizations are rarely indicative of RiPS in system operation plasmids, thus we are planning to also measure the fluorescence assays with the BioBricks on pSB3C5. In order to obtain sufficient BioBrick DNA the strains were grown in 250 mL LB with 100 μg/mL Ampicillin, awaiting plasmid isolation and plasmid swapping.
Emulsifier
The bricks for emulsifier production were assembled. The stock plasmids containing AlnA, OprG, R0011 and B0032 were digested and ligated into plasmid pSB1T3.
Digested were performed according to the digestion protocol:
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA | Needed fragment |
| 1 | 1.0 μg AlnA | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–AlnA–S’ |
| 2 | 1.0 μg OprG | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–OprG–S’ |
| 3 | 1.0 μg R0011 | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–R0011–S’ |
| 4 | 1.0 μg B0032 | XbaI | PstI | 2 (BioLabs) | ✓ | ‘X–B0032–P’ |
| 5 | 1.0 μg B0015 | XbaI | PstI | 2 (BioLabs) | ✓ | ‘X–B0015–P’ |
| 6 | 1.0 μg pSB1T3 | EcoRI | PstI | 2 (BioLabs) | ✓ | ‘E–linear pSB1T3–P’ |
The digestion products were ligated overnight:
| # | BioBrick | Fragment 1 | Fragment 2 | Recipient vector |
| 1 | K398202 | μL ‘E–R0011–S’ | μL ‘X–B0032–P’ | μL ‘E–linear pSB1T3–P’ |
| 2 | K398203 | μL ‘E–AlnA–S’ | μL ‘X–B0015–P’ | μL ‘E–linear pSB1T3–P’ |
| 3 | K398204 | μL ‘E–OprG–S’ | μL ‘X–B0015–P’ | μL ‘E–linear pSB1T3–P’ |
| 4 | negative control | - | - | μL ‘E–linear pSB1T3–P’ |
Lab work
Ordered DNA
We have now stock of the ordered DNA, to make a real BioBrick of this DNA we are going to ligated it into the iGEM plasmid backbone SB1C3. First we digested the plasmids:
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA | Needed fragment |
| 1 | 1 μg alkB2 | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–alkB2–P’ |
| 2 | 1 μg rubA3 | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–rubA3–P’ |
| 3 | 1 μg ladA | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–ladA–P’ |
| 4 | 1 μg ADH | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–ADH–P’ |
| 5 | 1 μg AlnA | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–AlnA–P’ |
| 6 | 1 μg OprG | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–OprG–P’ |
| 7 | 1 μg AlkS | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–AlkS–P’ |
| 8 | 1 μg PalkB | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–PalkB–P’ |
| 9 | 1 μg PalkS12 | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–PalkS12-P’ |
| 10 | 1 μg PhPFDα | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–PhPFDα–P’ |
| 11 | 1 μg PhPFDβ | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–PhPFDβ–P’ |
| 12 | 3 μg pSB1C3 | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–linear pSB1C3–P’ |
Solvent Tolerance and Hydrocarbon Sensing
Some BioBricks are in production. Digestion reaction
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA | Needed fragment |
| 1 | 1 μg PhPFDα | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–PhPFDα–S’ |
| 2 | 1 μg PhPFDβ | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–PhPFDβ–S’ |
| 3 | 1 μg AlkS | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–AlkS–S’ |
| 4 | 1 μg PalkB | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–PalkB–S’ |
| 5 | 1 μg B0032 | XbaI | PstI | 2 (BioLabs) | ✓ | ‘X–B0032–P’ |
| 6 | 1 μg B0015 | XbaI | PstI | 2 (BioLabs) | ✓ | ‘X–B0015–P’ |
| 7 | 1 μg E0422 | XbaI | PstI | 2 (BioLabs) | ✓ | ‘X–E0422–P’ |
| 8 | 3 μg pSB1T3 | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E-linear pSB1T3-P’ |
Emulsifier
Today the digestion products from yesterday were run on gel to see whether the plasmids were cut in the right way.
Lane description:
| # | Description | Expected Length (bp) | Status | Remarks |
| 1 | Biorad EZ marker | n/a | n/a | |
| 2 | Undigested pSB1T3 | 3507 | ✓ | |
| 3 | pSB1T3 + EcoRI + PstI | 1085, 2422 | ✓ | |
| 4 | Undigested AlnA | 3469 | ✓ | |
| 5 | AlnA + EcoRI + SpeI | 1071, 2398 | ✓ | |
| 6 | Undigested OprG | 3122 | ✓ | |
| 7 | OprG + EcoRI + SpeI | 708, 2414 | ✓ | |
| 8 | Undigested B0015 | 3318 | ✓ | |
| 9 | B0015 + XbaI + PstI | 155, 3163 | ✓ | fragment probably run of the gel |
| 10 | Undigested R0011 | 2134 | ✓ | Sample not fully loaded on gel |
| 11 | R0011 = EcoRI + SpeI | 78, 2056 | ? | fragment probably run of the gel |
| 12 | Undigested B0032 | 2092 | ✓ | |
| 13 | B0032 + XbaI + PstI | 39, 2053 | ? | fragment probably run of the gel |
| 14 | SmartLadder | n/a | n/a |
The ligation products that were incubated over night were transformed to Top10 competent cells according to the protocol.
Characterization of Anderson RBS sequences
The first attempt at measuring fluorescence was made in adherence with the [http://partsregistry.org/Part:BBa_K176011 protocol] proposed by the USTC team of 2009:
1. The overnight cultures of E.coli Top10 carrying K398500, K398501, K398502, K398503, K398504 and I13401 (LB, 37 ℃, 160 rpm) were measured for OD600 (see table below) and diluted 1:100.
| # | OD600 |
| K398500 | 1.443 |
| K398501 | 1.410 |
| K398502 | 1.448 |
| K398503 | 1.414 |
| K398504 | 1.552 |
| I13401 | 1.451 |
2. 100 μL and 300 μL aliquots of the diluted cultures were pipetted into 96-well plates in three-fold.
3. The plate was incubated at 37 ℃ while shaking for 3 hours.
4. The fluorescence and OD600 were measured for 16.30 hours with intervals 10 minutes by means of the Biotek Synergy plate reader with the excitation filter set at 485nm and the emission filter at 520nm. LB medium was used as blank reference.
Note: surprisingly no over night growth of the cultures was observed in M9 minimal medium containing glucose. This a larger volume of inoculate (in LB) was used for a second stab at over night growth in M9.
Plasmid purification of overnight cultures carrying K398500, K398501, K398502, K398503, K398504 in pSB1A3 was performed using the Qiagen Midi-prep plasmid isolation kit yielding sufficient plasmid DNA. These BioBricks can in future be placed under control of a low to medium copy number plasmid for comparison with high copy-number plasmid expression.
 "
"