Team:Stockholm/11 September 2010
From 2010.igem.org
Contents |
Andreas
Preparation of Top10 chemically competent cells
Picked 8 single colonies from the 10/9 LB plate with pipette tips. Colonies were restreaked onto an LB agar plate with Amp 100 to verify that they were not contaminated, and the pipette tips were then used to inoculate cultures of 5 ml LB. Cultures grown for 40 h in 30 °C w/o rotary shaking.
Extraction of RBS BioBrick (BBa_B0034)
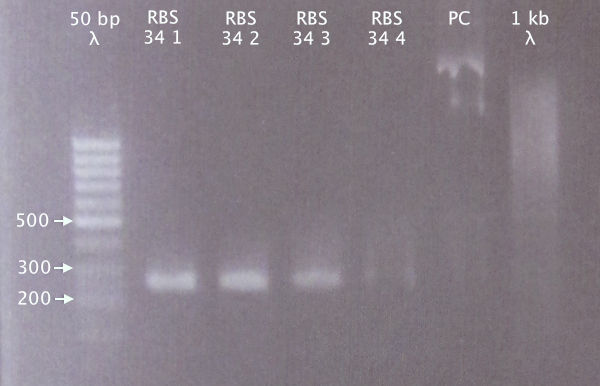
Colony PCR
Picked 4 colonies (RBS 1-4) from the 10/9 ON plate and ran a colony PCR.
- Positive control (PC): pSB1C3.RFP
- Standard colony PCR protocol
- Elongation: 0:45
Gel verification
1.5 % agarose, 90 V
Expected bands
- RBS 34: 250 bp
- PC: 1385 bp
Results
Relevant bands for clones 1-3. Weak band for clone 4. Selected clone 2 for plasmid prep.
ON cultures
Set double ON (until 13/9) cultures for RBS 34.
- 5 ml LB + Amp 100
- 37 °C, 80 rpm
- pSB1A2.RBS (BBa_B0034)
Extraction of RBS BioBrick (BBa_B0030)
Plasmid prep
From 10/9 ON culture
- Elution volume: 50 μl
- pSB1A2.RBS 30
Stored in -20 °C for later DNA conc. measurement.
Glycerol stock
- pSB1A2.BBa_B0030 (RBS) 2010-09-11
- Abbr.: pA.RBS 30
Cloning and site-directed mutagenesis of MITF
From Mimmi's 10/9 ON plate
Ran a PCR on four colonies from Mimmi's 10/9 transformation plate. Used original "pRc/CMV MITF -M" plasmid as control.
- m-MITF 1-4 11/9 (m-M 1-4)
- MITF plasmid (M)
PCR tubes
Standard colony PCR tubes (illustra Ready-to-Go PCR beads)
| PCR settings | ||
|---|---|---|
| 95 °C | 10:00 | |
| 95 °C | 0:30 | x5 |
| 55 °C | 0:30 | |
| 72 °C | 1:40 | |
| 95 °C | 0:30 | x25 |
| 72 °C | 2:10 | |
| 72 °C | 10:00 | |
PCR samples stored in 20 °C.
 |

|
 |

|
 |

|

|

|
 "
"