Project usu.html
From 2010.igem.org
Project
Contents |
Abstract
The future of synthetic biology lies in expanding our ability to engineer genes in new organisms. Our project develops a system to engineer the genome of the photosynthetic cyanobacterium Synechocystis sp. PCC6803, establishes expression standards for this species, and adds a set of characterized Synechocystis promoters and ribosome binding sites to the BioBrick toolbox. We developed a BioBrick vector that can be used to assemble parts and devices in E. coli. Upon transformation into Synechocystis, it integrates the device directly into the genome through homologous recombination. We utilized genes that were activated under a variety of conditions, from those responding to heat stress to ones oscillating under a circadian rhythm. The promoters and ribosome binding sites were converted into BioBrick-compatible parts, and subsequently characterized. Our success will enable the use of existing parts in new species, and will expand the range of devices that can be built.
Introduction
Synechocystis sp. PCC 6803
Synechocystis sp. PCC 6803 (hereafter referred to as PCC 6803) is a cyanobacterium, a photosynthetic bacterium, and one of the major model organisms in microbiology. The photosynthetic pathway of PCC 6803 has been extensively studied and its genome has been completely sequenced. It is capable of acting as both an autotroph, obtaining its energy from light and its carbon from the atmosphere, and as a heterotroph, using glycolysis of sugars as both an energy source and a carbon source. It also possesses the ability to uptake DNA present in its environment through natural transformation, making it a useful species for genetic manipulation (Zang, et al., 2007). PCC 6803 possesses many industrial applications including lipid recovery in biofuel production, PHB collection, and as a toxin biosensor (Wu, et al., 2001; Avramescu, et al., 1999). PCC 6803 also exhibits phototaxis and possesses an oscillating circadian clock, which may be of interest to genetic programmers. This species also has a set of well characterized secretion tags, which, unlike many species, are not sequence-dependent but rather charge-dependent, potentially allowing positively charged secretion tags from other species (such as E. coli) to be used in PCC 6803.
Project Justification
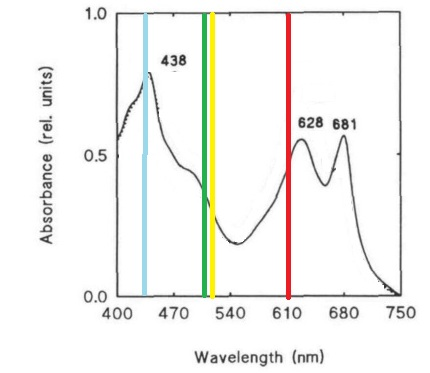
Despite the many potential uses and the promises of PCC 6803 in genetic research, new techniques for manipulating and studying bacterial genetics have not been adapted to this species. This project attempts the first step of the adaptations of the BioBrick biological part system to PCC 6803. This project seeks to create a BioBrick compatible integration vector for use in PCC 6803, since the species is more efficient at integrating genetic material into its chromosomes than maintaining separately replicating plasmids. This project also seeks to study the behaviors and strengths of 15 promoters from PCC 6803 which potentially are induced in light, darkness, heat stress, or nitrogen stress. Promoter strength and activation will be assessed using a cycle 3 mutant GFP (BBa_K20800), which has been used to test promoters in PCC 6803 in previous studies and is clearly distinguishable by spectrofluorometer despite the cell’s natural green color (Kunert, et al., 2000; Spence, et al., 2003). GFP was also chosen over CFP and RFP due to its excitation and emission wavelengths lying in a region where the photosynthetic system of PCC 6803 has reduced absorbance, which may improve accuracy of quantitative measurements by limiting interference (Figure 1) (Wilde, et al. 1995).
Figure 1. Absorbance spectrum of Synechocystis sp. PCC 6803 and the excitation and emission wavelengths of CFP, GFP, YFP, and RFP. Absorbance spectrum (black line) adaptated from Wilde, et al. (1995). Fluorescent proteins emission and excitation wavelengths shown as a single bar due to small relative distance between wavelengths, bars are color coded to the proteins.
Sigma Factors
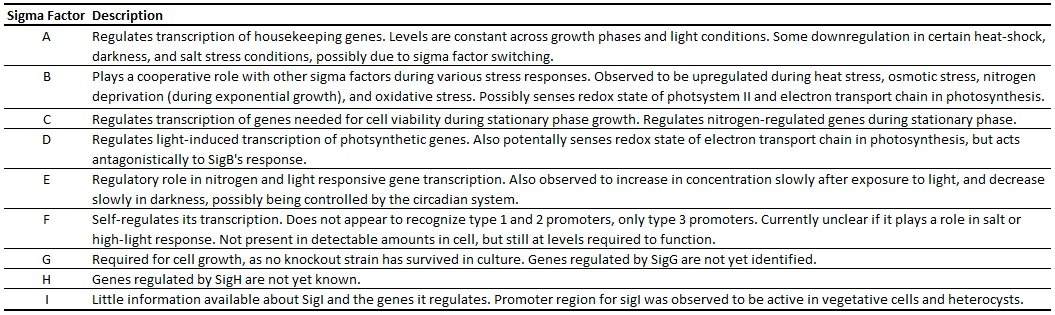
The transcription of genes in PCC 6803 is controlled in part by a system of nine currently known sigma factors (Table 1) (Imamura and Asayama, 2009). SigA controls the transcription of housekeeping genes, and its promoter will likely be a useful standard, as its concentration varies little over a range of temperatures, cell cycle phases, and stress conditions. SigE is upregulated by light and downregulated in the dark, potentially contributing to circadian rhythm-based gene transcription. The other sigma factors are detailed in Table 1 and are thoroughly reviewed in Imamura and Asayama (2009).
Table 1. Sigma Factors in Synechocystis sp. PCC 6803. Potential functions of genes listed, if known. Adapted from Imamura and Asayama (2009).
Promoter Types
These sigma factors act on three classes of promoter sequences (summarized from Imamura and Asayama, 2009):
- Type I promoters have the typical prokaryotic structure of -10 and -30 regions, and are generally activated by SigA. Type I promoters have high homology to E. coli’s sigma70 binding sequence, and could potentially be active in both species.
- Type II promoters have only the -10 region, but may include enhancer sequences for stress response.
- Type III promoters have -12 and -32 regions, are activated through SigF, and are transcriptionally independent of the other two types
References and Useful Papers
Avramescu A, Rouillon R, and Carpentier R. 1999. Potential for use of a cyanobacterium Synechocystis sp. immobilized in poly(vinylalcohol): Application to the detection of pollutants. Biotechnology Techniques 13: 559-562.
Haring V, Scholz P, Scherzinger E, Frey J, Derbyshire K, Hatfull G, Willetts NS, and Bagdasarian M. 1985. Protein RepC is involved in copy number control of the broad host range plasmid RSF1010. Proceedings of the National Academy of Sciences of the United States of America 82: 6090-6094.
Imamura S and Asayama M. 2009. Sigma Factors for Cyanobacterial Transcription. Gene Regulation and Systems Biology 3: 65-87.
Imamura S, Asayama M, Takahashi H, Tanaka K, Takahashi H, and Shirai M. 2003. Antagonistic dark/light-induced SigB/SigD, group 2 sigma factors, expression through redox potential and their roles in cyanobacteria. FEBS Letters 554: 357-362.
Imamura S, Tanaka K, Shirai M, and Asayama M. 2006. Growth phase-dependent activation of nitrogen-related genes by a control network of group 1 and group 2 sigma factors in a cyanobacterium. Journal of Biological Chemistry 281: 2668–2675.
Kunert A, Hagemann M, and Erdmann N. 2000. Construction of promoter probe vectors for Synechocystis sp. PCC 6803 using the light-emitting reporter systems GFP and LuxAB. Journal of Microbiological Methods 41: 185-194.
Ludwig A, Heimbucher T, Gregor W, Czerny T, and Schmetterer G. 2008. Transformation and gene replacement in the facultatively chemoheterotrophic, unicellular cyanobacterium Synechocystis sp. PCC6714 by electroporation. Applied Microbiology and Biotechnology 78: 729-735.
Singh AK, Summerfield TC, Li H, an d Sherman LA. 2006. The heat shock response in the cyanobacterium Synechocystis sp. Strain PCC 6803 and regulation of gene expression by HrcA and SigB. Archives of Microbiology 186: 273-286.
Spence E, Sarcina M, Ray N, Moller SG, Mullineaux CW, and Robinson C. 2003. Membrane-specific targeting of green fluorescent protein by the Tat pathway in the cyanobacterium Synechocystis PCC 6803. Molecular Microbiology 48: 1481-1489.
Wilde A, Härtel H, Hübschmann T, Hoffmann P, Shestakov SV, and Börner T. 1995. Inactivation of a Synechocystis sp strain PCC 6803 gene with homology to conserved chloroplast open reading frame 184 increases the photosystem II-to-photosystem I ratio. The Plant Cell 7: 649-658.
Wu GF, Wu QY, and Shen ZY. 2001. Accumulation of poly-[beta]-hydroxybutyrate in cyanobacterium Synechocystis sp. PCC6803. Bioresource Technology 76: 85-90.
Zang X, Liu B, Liu S, Arunakumara KK, and Zhang X. 2007. Optimum conditions for transformation of Synechocystis sp. PCC 6803. Journal of Microbiology (Seoul, Korea) 45: 241-245
 "
"