Modeling page toggle here
From 2010.igem.org
Modeling Details of Toggle Switch
Key points of this modeling are:
- LacI protein tends to form LacI dimer, which tends to form LacI tetramer further.
- Only LacI tetramer binds with pLacI gene and thus repressing expression of its downstream genes.
- TetR protein only forms dimer.
- Only tetR dimer binds with pTet gene and thus repressing expression of its downstream genes.
- One IPTG molecule binds with one LacI tetramer to form complex IPTG:LacI4 and one aTc molecule binds with one tetR dimer to form complex aTc:TetR2.
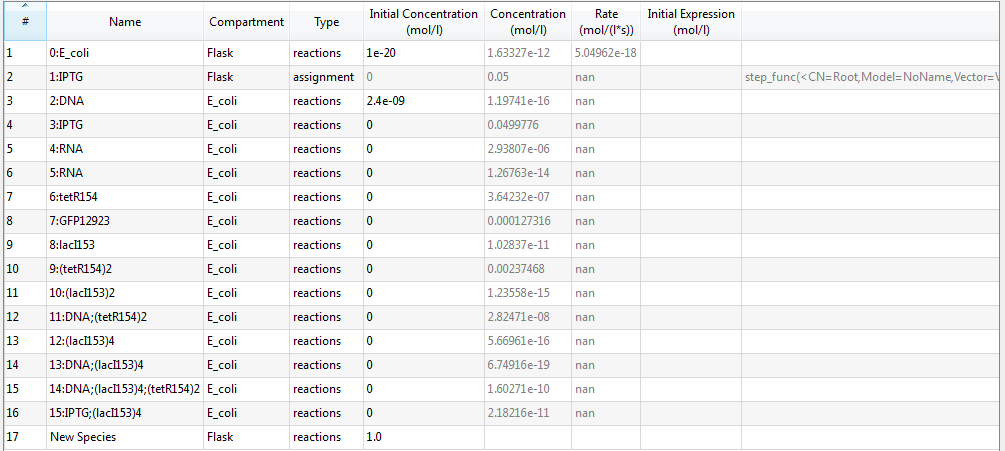
An overview of species list is provided:
where each species with its specific identifier has its own meaning:
- 0:E_coli : E.coli cell;
- 1:IPTG: IPTG in flask;
- 2:DNA: initial transformed plasmids;
- 3:IPTG: IPTG in E.coli due to diffusion;
- 4:RNA: mRNA of tetR and GFP;
- 5:RNA: mRNA of LacI;
- 6:tetR154: tetR protein;
- 7:GFP12923: GFP;
- 8:lacI153: LacI protein;
- 9:(tetR154)2: tetR dimer;
- 10:(lacI153)2: LacI dimer;
- 11:DNA;(tetR154)2: complex of tetR dimer binding to pTet gene of plasmid DNA;
- 12:(lacI153)4: LacI tetramer;
- 13:DNA;(lacI153)4: complex of LacI tetramer binding to pLacI gene of plasmid DNA;
- 14:DNA;(lacI153)4;(tetR154)2: complex of plasmid DNA with both pLacI and pTet genes bound with LacI tetramer and tetR dimer, respectively;
- 15:IPTG;(lacI153)4: complex of IPTG binding with LacI tetramer;
Since there are 65 reactions in total, we only provide a screenshot (we strongly recommend users to run our example to learn more details about automatic modeling):
Moreover, parameters of our modeling network are provided (we spend much efforts on tuning parameters) based on papers and estimates together:
- Initial time: 0 s
- Initial volumes:
- Flask: 0.4 l
- E_coli: 1.6856e-012 l
- Initial concentrations:
- 0:E_coli: 1e-020 mol/l
- 1:IPTG: 0 mol/l
- 2:DNA: 2.4e-009 mol/l
- 3:IPTG: 0 mol/l
- 4:RNA: 0 mol/l
- 5:RNA: 0 mol/l
- 6:tetR154: 0 mol/l
- 7:GFP12923: 0 mol/l
- 8:lacI153: 0 mol/l
- 9:(tetR154)2: 0 mol/l
- 10:(lacI153)2: 0 mol/l
- 11:DNA;(tetR154)2: 0 mol/l
- 12:(lacI153)4: 0 mol/l
- 13:DNA;(lacI153)4: 0 mol/l
- 14:DNA;(lacI153)4;(tetR154)2: 0 mol/l
- 15:IPTG;(lacI153)4: 0 mol/l
- Reaction parameters:
- reproduction of Ecoli cell
- growth constant: 0.000192 1/s
- maximum concentration: 1.66e-012 mol/l
- diffusion of IPTG molecule
- diffusion constant: 0.014 1/s
- transcription
- transcription rate of pTet/pLacI: 0.5 1/s
- replication of reverse pSB3C5
- replication constant: 0.003 1/s
- repression of reverse pSB3C5 replication
- max concentration of pSB3C5: 2.847e-008 mol/l
- translation
- translation rate: 0.1 1/s
- degradation of mRNAs
- degradation of mRNA molecules: 0.0048 1/s
- degradation of proteins in E.coli_3
- degradation rate of proteins: 0.0023 1/s
- Laci-Laci dimerization
- kon: 1.25e+007 l/(mol*s)
- koff: 10 1/s
- binding forward-ptetR:tetR2
- kon: 1e+008 l/(mol*s)
- koff: 0.001 1/s
- tetR-tetR dimerization_2
- kon: 4e+011 1/(mol*s)
- koff: 10 1/s
- LacI2-LacI2 dimerization
- kon: 1e+014 l/(mol*s)
- koff: 10 1/s
- binding reverse-placI:lacI4
- kon: 4e+011 l/(mol*s)
- koff: 0.04 1/s
- IPTG:lacI4 binding
- kon: 154000 l/(mol*s)
- koff: 0.2 1/s
- reproduction of Ecoli cell
 "
"