Team:Wisconsin-Madison/experiments
From 2010.igem.org
| (36 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
#nav_content { | #nav_content { | ||
float: left; | float: left; | ||
| - | width: | + | position: fixed; |
| + | width: 230px; | ||
margin-left: 0px; | margin-left: 0px; | ||
| - | margin-right: | + | margin-right: 0px; |
| - | margin-top: | + | margin-top: 5px; |
| - | height: | + | height: 280px; |
| + | } | ||
| + | #page_content { | ||
| + | float: left; | ||
| + | width: 690px; | ||
| + | text-align: justify; | ||
| + | margin-left: 230px; | ||
| + | margin-right: auto; | ||
| + | padding-left: 0px; | ||
| + | padding-right: 0px; | ||
| + | position: relative; | ||
} | } | ||
</style> | </style> | ||
| Line 14: | Line 25: | ||
</html>__TOC__<html> | </html>__TOC__<html> | ||
</div> | </div> | ||
| + | <div id="page_content"> | ||
</html> | </html> | ||
| - | + | ==Encapsulation== | |
| - | + | ===Colonic Acid Quantification=== | |
| - | === | + | Download procedure [[Media:UW_colonicacidassay2.pdf|here]] |
| + | |||
{| border="1" cellpadding="5" cellspacing="0" align="center" | {| border="1" cellpadding="5" cellspacing="0" align="center" | ||
|+ align="center" style="color:#000000;" |''Parts used in this experiment'' | |+ align="center" style="color:#000000;" |''Parts used in this experiment'' | ||
| Line 27: | Line 40: | ||
|- | |- | ||
|<partinfo>BBa_k318500</partinfo> | |<partinfo>BBa_k318500</partinfo> | ||
| - | |Produces Trascription Factor | + | |Produces Trascription Factor RcsA |
|Inducible - IPTG | |Inducible - IPTG | ||
| - | | | + | |[[media:BBa_K318500_pSB1C3.zip|here]] |
|- | |- | ||
|<partinfo>BBa_k318501</partinfo> | |<partinfo>BBa_k318501</partinfo> | ||
| - | |Produces Trascription Factor | + | |Produces Trascription Factor RcsB |
|Inducible - IPTG | |Inducible - IPTG | ||
| - | | | + | |[[media:BBa_K318501_pSB1C3.zip|here]] |
|- | |- | ||
|<partinfo>BBa_k318502</partinfo> | |<partinfo>BBa_k318502</partinfo> | ||
|Produces Trascription Factor RcsA & RcsB | |Produces Trascription Factor RcsA & RcsB | ||
|Inducible - IPTG | |Inducible - IPTG | ||
| - | | | + | |[[media:BBa_K318502_pSB1C3.zip|here]] |
| + | |- | ||
| + | |<partinfo>BBa_k200021</partinfo> | ||
| + | |Empty Vector/Contol | ||
| + | |Inducible - IPTG | ||
| + | |NA | ||
|} | |} | ||
| + | <br> | ||
| + | '''Background''' | ||
| + | Colonic Acid is a polysaccharide containing a repeat unit with D-glucose, L-fucose, D-galactose, and D-glucuronate. Biological extracts often contain compounds, which under heating with H2SO4 yield brown products absorbing between 396 nm and 427 nm. Colonic acid can be estimated by measuring L-fucose content. | ||
| + | <br> | ||
| + | '''Materials''' | ||
| + | *H2SO4/H2O (6:1 v/v) | ||
| + | *Cysteine hydrochloride | ||
| + | *L-Frucose (calibration curve) | ||
| + | <br> | ||
| + | '''Cell Culture:''' (Final 50ml sample) | ||
| + | #Transform plasmids for high protein production | ||
| + | #Incubate plate overnight (do not place in fridge before next step) | ||
| + | #Prepare liquid culture of each sample | ||
| + | #Induce with 1mM IPTG at OD600 = 0.05 | ||
| + | #Grow cells to stationary phase | ||
| + | <br> | ||
| + | '''Protocol:''' | ||
| + | #Prepare overnight liquid culture of 50 ml sample | ||
| + | #Measure OD600 | ||
| + | #To inactivate EPS-degrading enzymes and completely release EPS from cell surface: | ||
| + | ##Boil sample for 15 min | ||
| + | ##Cool to room temp | ||
| + | ##Centrifuge at 14,000g for 30 min at 4°C | ||
| + | #Add three volumes of 70% ethanol to 40 ml of supernatant fraction | ||
| + | #Place in 4°C overnight | ||
| + | #Centrifuge at 14,000g for 30 min at 4°C | ||
| + | #Dissolve pellet in 1 ml of sterile distilled water | ||
| + | <br> | ||
| + | '''Quantification:''' Use negative controls of glucose and sterile distilled water | ||
| + | #Mix 4.5 ml of H2SO4/H2O (6:1 v/v) with 100uL of 1ml sample | ||
| + | #Heat mixture to 100°C for 20 min | ||
| + | #Cool to room temperature | ||
| + | #Measure absorbance at 396 nm and 427 nm | ||
| + | #Add 100 μL of 10M cysteine hydrochloride | ||
| + | #Measure absorbance at 396 nm and 427 nm | ||
| + | #Difference in these measurements (after subtracted from pre-cysteine addition absorbance) can be directly correlated to methylpentose concentration by using a standard curve obtained with a fucose concentration ranging from 5 μg/ml to 100 μg/ml | ||
| + | <br> | ||
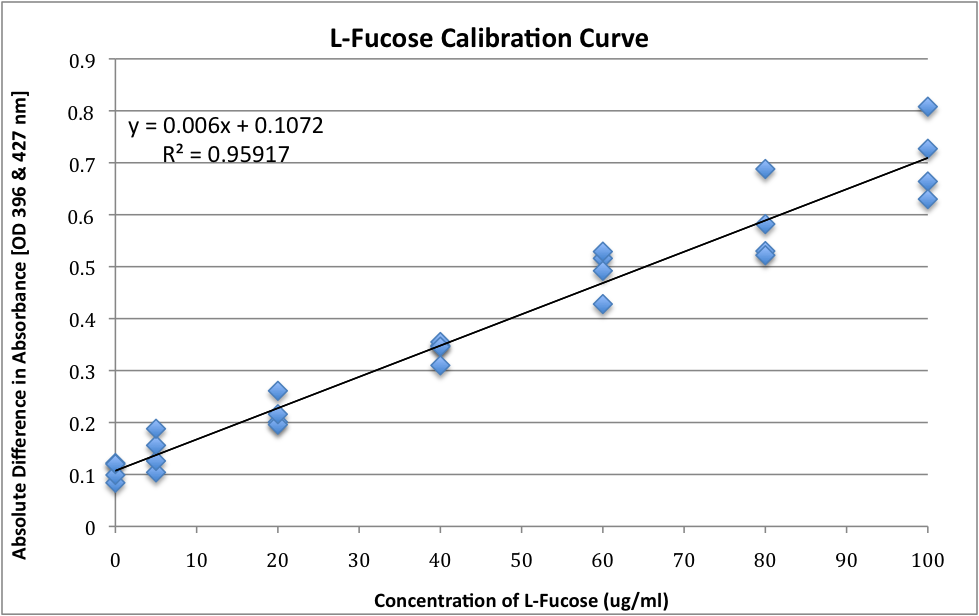
| + | '''L-Fucose – Standard Curve''' | ||
| + | *Concentrations of L-Fucose ranging from 0 μg/ml to 100 μg/ml | ||
| + | *L-Fucose was added to the appropriate amount of sterile distilled water to result in the following concentrations and 1ml of this preparation was taken from step 8(b) to 8(h) from Colonic Acid Quantification Assay (above) | ||
| + | <br> | ||
| + | [[Image:UW_LFucoseCurve.png|400px]] | ||
| - | + | References: [[http://www.nature.com/ismej/journal/v2/n6/full/ismej200824a.html Link1]],[[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WK7-4MMWHP9-1&_user=217827&_coverDate=03%2F16%2F2007&_rdoc=1&_fmt=full&_orig=search&_cdi=6899&_sort=d&_docanchor=&view=c&_acct=C000011279&_version=1&_urlVersion=0&_userid=217827&md5=98bee864d6a8931008db26c024e105e0#secx10 Link2]][[http://pubs.acs.org/doi/abs/10.1021/ja01129a015 Link3]] | |
| - | + | <br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | # | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Cell Survivability Testing=== | |
| - | + | {| border="1" cellpadding="5" cellspacing="0" align="center" font-size="80%" | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | {| border="1" cellpadding="5" cellspacing="0" align="center" | + | |
|+ align="center" style="color:#000000;" |''Parts used in this experiment'' | |+ align="center" style="color:#000000;" |''Parts used in this experiment'' | ||
|- | |- | ||
| Line 76: | Line 116: | ||
|- | |- | ||
|<partinfo>BBa_k318500</partinfo> | |<partinfo>BBa_k318500</partinfo> | ||
| - | |Produces Trascription Factor | + | |Produces Trascription Factor RcsA |
|Inducible - IPTG | |Inducible - IPTG | ||
| - | | | + | |[[media:BBa_K318500_pSB1C3.zip|here]] |
|- | |- | ||
|<partinfo>BBa_k318501</partinfo> | |<partinfo>BBa_k318501</partinfo> | ||
| - | |Produces Trascription Factor | + | |Produces Trascription Factor RcsB |
|Inducible - IPTG | |Inducible - IPTG | ||
| - | | | + | |[[media:BBa_K318501_pSB1C3.zip|here]] |
|- | |- | ||
|<partinfo>BBa_k318502</partinfo> | |<partinfo>BBa_k318502</partinfo> | ||
|Produces Trascription Factor RcsA & RcsB | |Produces Trascription Factor RcsA & RcsB | ||
|Inducible - IPTG | |Inducible - IPTG | ||
| - | | | + | |[[media:BBa_K318502_pSB1C3.zip|here]] |
| + | |- | ||
| + | |<partinfo>BBa_k200021</partinfo> | ||
| + | |Empty Vector/Contol | ||
| + | |Inducible - IPTG | ||
| + | |NA | ||
|} | |} | ||
| - | |||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | [[Media: | + | Download procedure [[Media:UW_Cell_Survivability_Protocol.pdf|here]] |
| - | [[ | + | '''Materials''' |
| + | |||
| + | *1M HCl | ||
| + | *1M NaOH | ||
| + | *LB | ||
| + | *1M IPTG | ||
| + | |||
| + | '''Protocol for Acid Survivability Test''' | ||
| + | |||
| + | # Grow cells overnight in LB (RcsA-MG1655(A), RcsB-MG1655(B), RcsA+RcsB-MG1655(AB), and MG1655 with empty vector K200021(C)) | ||
| + | # Dilute samples to OD600 = 0.05 and induce with 1mM IPTG | ||
| + | # Harvest cells at OD between 1 to 2, Wait until the slowest one is at OD=1.5 | ||
| + | # Make LB+ITPG cultures that are OD=1.5, and the volume should between 1ml to 5ml. | ||
| + | # Spin down the cells at 3000g for 2-3mins. | ||
| + | # Re-suspend the cells in pH LB medium (pH=2, 4, 7) | ||
| + | # Plate cell samples at time zero. | ||
| + | # Place all the samples into the 37C shaker and let them grow for 4hs. | ||
| + | # Remove the samples from the shaker, and record the OD. | ||
| + | # Make a series dilution for each sample. (10E-3, 10E-4, 10E-5, 10E-6, 10E-7, 10E-8, 10E-9) | ||
| + | # Plate 20ul of each dilution of each sample on a CM mini-Petridish plate. | ||
| + | # Put all the plates in to 37C incubator and let the cells grow overnight. | ||
| + | |||
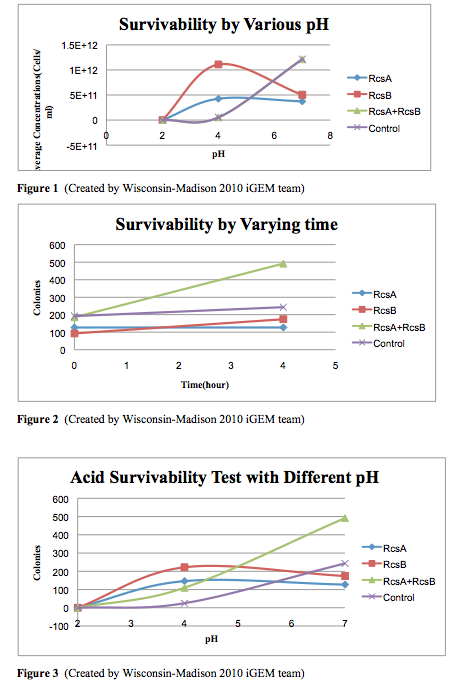
| + | [[Image:Cell Survivability Testing Graphs.jpg]] | ||
| + | |||
| + | ==Inducible-Repressible Expression== | ||
| + | ===Characterize pH Promoter=== | ||
| + | |||
| + | Download testing procedure (.pdf) [[Media:GadAp_Test_Protocol.pdf|here]]. | ||
| + | |||
| + | Download example spreadsheet (.xls) [[Media:Copy of Final Friday Eval.xls|here]]. | ||
| + | |||
| + | {| border="1" cellpadding="5" cellspacing="0" align="center" | ||
| + | |+ align="center" style="color:#000000;" |''Parts used in this experiment'' | ||
| + | |- | ||
| + | |'''Part Number''' | ||
| + | |'''Function''' | ||
| + | |'''Induction''' | ||
| + | |'''Zip File''' | ||
| + | |- | ||
| + | |<partinfo>BBa_k318513</partinfo> | ||
| + | |Produces RFP | ||
| + | |stationary phase | ||
| + | |[[media:BBa_K318512_pSB1C3.zip|here]] | ||
| + | |} | ||
| + | <html> | ||
| + | <div> | ||
| + | </html> | ||
Latest revision as of 03:22, 28 October 2010
Encapsulation
Colonic Acid Quantification
Download procedure here
| Part Number | Function | Expression Type | Zip File |
| <partinfo>BBa_k318500</partinfo> | Produces Trascription Factor RcsA | Inducible - IPTG | here |
| <partinfo>BBa_k318501</partinfo> | Produces Trascription Factor RcsB | Inducible - IPTG | here |
| <partinfo>BBa_k318502</partinfo> | Produces Trascription Factor RcsA & RcsB | Inducible - IPTG | here |
| <partinfo>BBa_k200021</partinfo> | Empty Vector/Contol | Inducible - IPTG | NA |
Background
Colonic Acid is a polysaccharide containing a repeat unit with D-glucose, L-fucose, D-galactose, and D-glucuronate. Biological extracts often contain compounds, which under heating with H2SO4 yield brown products absorbing between 396 nm and 427 nm. Colonic acid can be estimated by measuring L-fucose content.
Materials
- H2SO4/H2O (6:1 v/v)
- Cysteine hydrochloride
- L-Frucose (calibration curve)
Cell Culture: (Final 50ml sample)
- Transform plasmids for high protein production
- Incubate plate overnight (do not place in fridge before next step)
- Prepare liquid culture of each sample
- Induce with 1mM IPTG at OD600 = 0.05
- Grow cells to stationary phase
Protocol:
- Prepare overnight liquid culture of 50 ml sample
- Measure OD600
- To inactivate EPS-degrading enzymes and completely release EPS from cell surface:
- Boil sample for 15 min
- Cool to room temp
- Centrifuge at 14,000g for 30 min at 4°C
- Add three volumes of 70% ethanol to 40 ml of supernatant fraction
- Place in 4°C overnight
- Centrifuge at 14,000g for 30 min at 4°C
- Dissolve pellet in 1 ml of sterile distilled water
Quantification: Use negative controls of glucose and sterile distilled water
- Mix 4.5 ml of H2SO4/H2O (6:1 v/v) with 100uL of 1ml sample
- Heat mixture to 100°C for 20 min
- Cool to room temperature
- Measure absorbance at 396 nm and 427 nm
- Add 100 μL of 10M cysteine hydrochloride
- Measure absorbance at 396 nm and 427 nm
- Difference in these measurements (after subtracted from pre-cysteine addition absorbance) can be directly correlated to methylpentose concentration by using a standard curve obtained with a fucose concentration ranging from 5 μg/ml to 100 μg/ml
L-Fucose – Standard Curve
- Concentrations of L-Fucose ranging from 0 μg/ml to 100 μg/ml
- L-Fucose was added to the appropriate amount of sterile distilled water to result in the following concentrations and 1ml of this preparation was taken from step 8(b) to 8(h) from Colonic Acid Quantification Assay (above)
References: http://www.nature.com/ismej/journal/v2/n6/full/ismej200824a.html Link1,[[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WK7-4MMWHP9-1&_user=217827&_coverDate=03%2F16%2F2007&_rdoc=1&_fmt=full&_orig=search&_cdi=6899&_sort=d&_docanchor=&view=c&_acct=C000011279&_version=1&_urlVersion=0&_userid=217827&md5=98bee864d6a8931008db26c024e105e0#secx10 Link2]]http://pubs.acs.org/doi/abs/10.1021/ja01129a015 Link3
Cell Survivability Testing
| Part Number | Function | Expression Type | Zip File |
| <partinfo>BBa_k318500</partinfo> | Produces Trascription Factor RcsA | Inducible - IPTG | here |
| <partinfo>BBa_k318501</partinfo> | Produces Trascription Factor RcsB | Inducible - IPTG | here |
| <partinfo>BBa_k318502</partinfo> | Produces Trascription Factor RcsA & RcsB | Inducible - IPTG | here |
| <partinfo>BBa_k200021</partinfo> | Empty Vector/Contol | Inducible - IPTG | NA |
Download procedure here
Materials
- 1M HCl
- 1M NaOH
- LB
- 1M IPTG
Protocol for Acid Survivability Test
- Grow cells overnight in LB (RcsA-MG1655(A), RcsB-MG1655(B), RcsA+RcsB-MG1655(AB), and MG1655 with empty vector K200021(C))
- Dilute samples to OD600 = 0.05 and induce with 1mM IPTG
- Harvest cells at OD between 1 to 2, Wait until the slowest one is at OD=1.5
- Make LB+ITPG cultures that are OD=1.5, and the volume should between 1ml to 5ml.
- Spin down the cells at 3000g for 2-3mins.
- Re-suspend the cells in pH LB medium (pH=2, 4, 7)
- Plate cell samples at time zero.
- Place all the samples into the 37C shaker and let them grow for 4hs.
- Remove the samples from the shaker, and record the OD.
- Make a series dilution for each sample. (10E-3, 10E-4, 10E-5, 10E-6, 10E-7, 10E-8, 10E-9)
- Plate 20ul of each dilution of each sample on a CM mini-Petridish plate.
- Put all the plates in to 37C incubator and let the cells grow overnight.
Inducible-Repressible Expression
Characterize pH Promoter
Download testing procedure (.pdf) here.
Download example spreadsheet (.xls) here.
| Part Number | Function | Induction | Zip File |
| <partinfo>BBa_k318513</partinfo> | Produces RFP | stationary phase | here |
 "
"