Team:WITS-South Africa/e-chromi
From 2010.igem.org

E.chromi Orange
Our Lacto-Report construct design included an E.chromi Biobrick, as designed by Cambridge's 2009 iGEM team. The Biobrick that we initially selected was Orange E.chromi (BB_k2700).
However, after a number of attempts to assemble this part into our machine using the Standard Assembly method with no success, we decided to determine if all the correct Biobrick restriction sites were present. We also suspected that perhaps one of the sites had been methylated, so we transformed the initial Biobrick plasmid received from the DNA distribution into a methylase negative strain of E.coli (Sure cells). We then prepared plasmid from one of the colonies and performed a series of digests on the plasmid.
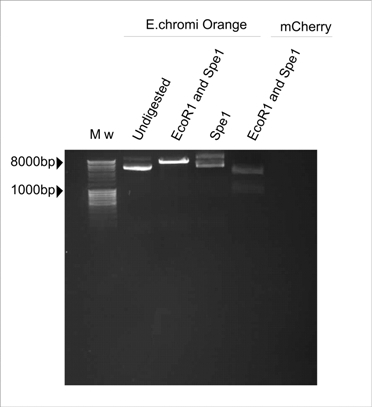
Figure 1. E.chromi Orange digested with various restriction enzymes to determine the presence of the Biobrick restriction sites.
In Figure 1, the plasmid sample digested with Spe1 (Fermentas) resembles the undigested control. The plasmid sample double-digested with EcoR1 and Spe1 (Fermentas) is linearised by cutting with EcoR1 but there are no distinct insert and plasmid bands, which is the banding pattern we expect to see after digesting with these two enzymes. The expected pattern is clearly visible in the mCherry control which was also digested with EcoR1 and Spe1 (Fermentas).
E.chromi Dark Green
We then decided to try and use the Dark Green E.chromi biobrick (Bb_K). Once again, although the part was excised using EcoR1 and Spe1, when we attempted to clone anything in front of this part, by digestion with EcoR1 and Xba1, we could not obtain any positive clones.
Suspecting that perhaps the restriction sites were not present, we tested the plasmid samples.
[[image: wits_xbal_site_
 "
"