Team:WITS-South Africa/Machine Design
From 2010.igem.org
(→Machine Testing) |
(→Machine Testing) |
||
| Line 41: | Line 41: | ||
===Machine Testing=== | ===Machine Testing=== | ||
<br /> | <br /> | ||
| - | A culture of 2ml of ''E. coli''-containing Lacto-detect was grown overnight at | + | A culture of 2ml of ''E. coli''-containing Lacto-detect was grown overnight at 37C and then transfered to 25 ml of ampicillin LB broth. This innoculent was grown at 37C in a shaking incubator for 2.5 hours so as to ensure that the bacteria were in their exponential growth phase. A baseline reading (fig 1) was taken at this point (tile A), after which 10% 1mM IPTG was added and then returned to the shaking incubator. An aliquot was then imaged after 30min (tile B) and again after 1 hour (tile C). All aliquots used in imaging were 100 ul. As is seen in the baseline image (tile A) the degree of fluorescent activation is minimal before the addition and incubation of IPTG. Tile B shows the fluorescent activation after 30min incubation with IPTG - there is very little visible fluorescent activation. After an hour of incubation (tile C) there is a marked increase in the degree of fluorescence thus illustrating the time taken for IPTG to induce a response. <br /> <br /> |
| - | [[Image:Montage_of_M1I_d.jpg|850px]] <br /> <br /> <br /> <br /> | + | [[Image:Montage_of_M1I_d.jpg|850px]] <br /> |
| + | Figure 1 <br /> <br /> <br /> | ||
| - | Once the induction of Lacto-detect had reached an appreciable level, it was added to an aliquot of ''B. subtilis'' containing a construct driven by the PlcR promoter. Baseline images of both Lacto-detect (tile A) and the ''B. subtilis'' population (tile B) are given as well as a combined baseline image (tile C). <br /> <br /> | + | Once the induction of Lacto-detect had reached an appreciable level, it was added to an aliquot of ''B. subtilis'' containing a construct driven by the PlcR promoter. Baseline images (fig 2) of both Lacto-detect (tile A) and the ''B. subtilis'' population (tile B) are given as well as a combined baseline image (tile C). <br /> <br /> |
[[Image:Montage_of_Mixed.jpg|850px]] <br /> | [[Image:Montage_of_Mixed.jpg|850px]] <br /> | ||
| + | Figure 2 <br /> <br /> <br /> | ||
| + | |||
| + | Once the induced aliquot of Lacto-detect was added, the combined sample was imaged at 30min intervals for 6hours so as to show the activation of the PlcR-driven ''B. subtilis'' population (figure 3). Images at each epoch were taken using both Venus and mCherry-specific filters and are depicted as alphabetical sets - A&B, C&D etc. Taking particular note of the image set M&N (3.5h), there is marked increase in mCherry fluoresence. This is indicative of the active communication between Lacto-detect and the PlcR ''B. subtilis'' population, thus illustrating the efficacy of the PlcR-PapR quorum network. <br /> <br /> | ||
| + | [[Image:Montage_of_Combo.jpg|850px]] <br /> Figure 3 | ||
| + | |||
| + | Figure 4 shows both populations of | ||
Revision as of 14:50, 27 October 2010

Contents |
Lacto-detect
Purpose:
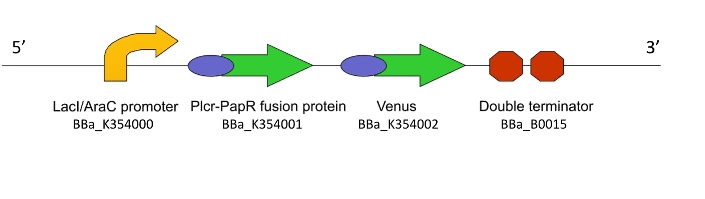
a) To act as the ‘Detector’ Machine within the population and produce the quorum signalling peptide in response to an input signal
So how did we end up selecting the final parts of our Machines? Each one was chosen after much consideration and scouring of the literature to select the most suitable biological system. For a design rationale of why we selected the parts that we did to create this machine, click here
For an account of how we assemblied and verified Lacto-detect, click here
Lacto-report
For a design rationale of why we selected the parts that we did to create this machine, click here
We were not able to finish constructing Lacto-report in it's entirety, due to time constraints and issues with the E.chromi Biobrick parts. (For more details, click here.)
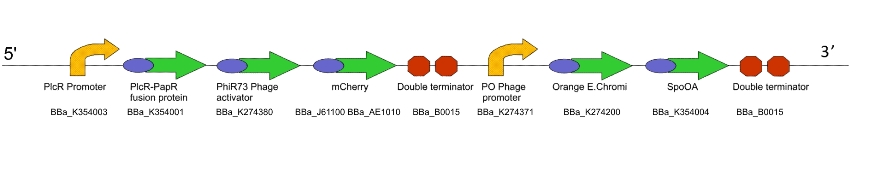
For testing and characterising the behaviour of the quorum sensing mechanism in a model Gram-positive bacillus, we used an intermediate machine construct we dubbed Lacto-
Lacto-test
Purpose:
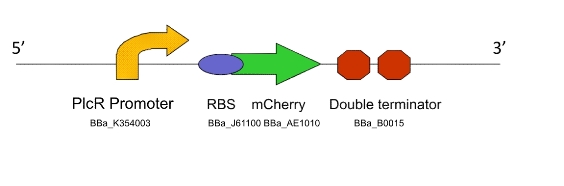
a) To show that the PlcR promoter is activated in L. gasseri by measuring fluorescence after the addition of exogenous PlcR and PapR proteins
Machine Testing
A culture of 2ml of E. coli-containing Lacto-detect was grown overnight at 37C and then transfered to 25 ml of ampicillin LB broth. This innoculent was grown at 37C in a shaking incubator for 2.5 hours so as to ensure that the bacteria were in their exponential growth phase. A baseline reading (fig 1) was taken at this point (tile A), after which 10% 1mM IPTG was added and then returned to the shaking incubator. An aliquot was then imaged after 30min (tile B) and again after 1 hour (tile C). All aliquots used in imaging were 100 ul. As is seen in the baseline image (tile A) the degree of fluorescent activation is minimal before the addition and incubation of IPTG. Tile B shows the fluorescent activation after 30min incubation with IPTG - there is very little visible fluorescent activation. After an hour of incubation (tile C) there is a marked increase in the degree of fluorescence thus illustrating the time taken for IPTG to induce a response.
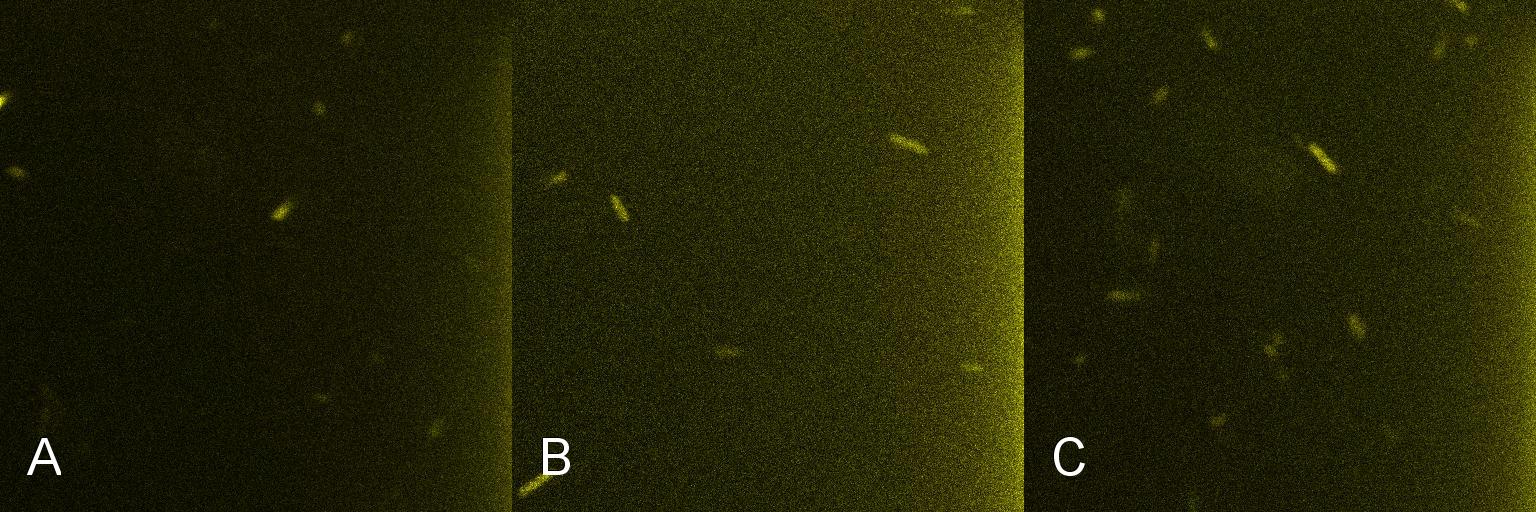
Figure 1
Once the induction of Lacto-detect had reached an appreciable level, it was added to an aliquot of B. subtilis containing a construct driven by the PlcR promoter. Baseline images (fig 2) of both Lacto-detect (tile A) and the B. subtilis population (tile B) are given as well as a combined baseline image (tile C).
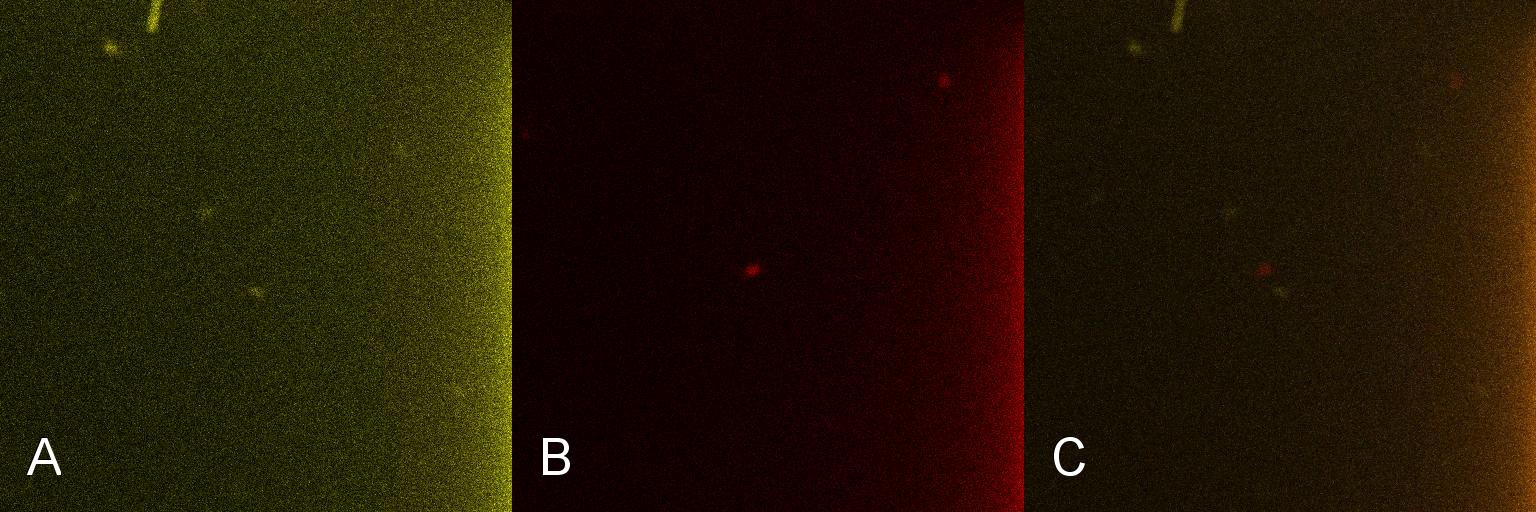
Figure 2
Once the induced aliquot of Lacto-detect was added, the combined sample was imaged at 30min intervals for 6hours so as to show the activation of the PlcR-driven B. subtilis population (figure 3). Images at each epoch were taken using both Venus and mCherry-specific filters and are depicted as alphabetical sets - A&B, C&D etc. Taking particular note of the image set M&N (3.5h), there is marked increase in mCherry fluoresence. This is indicative of the active communication between Lacto-detect and the PlcR B. subtilis population, thus illustrating the efficacy of the PlcR-PapR quorum network.
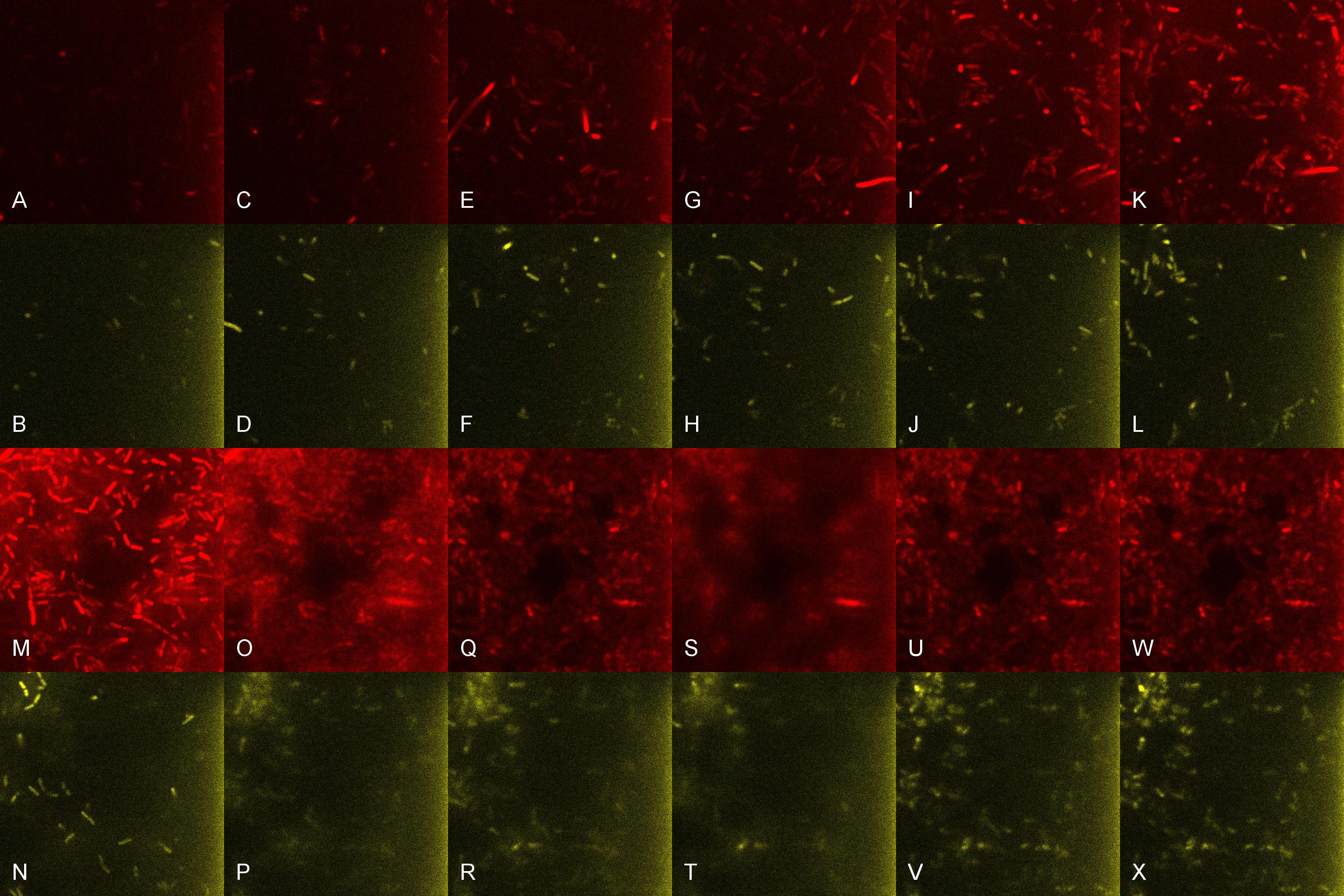
Figure 3
 "
"