Team:WITS-South Africa/Machine Design
From 2010.igem.org
| (29 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{{Template:WITS-South_Africa_Main_Menu_0910}} | {{Template:WITS-South_Africa_Main_Menu_0910}} | ||
| - | = | + | <div style="padding:40px;"> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ==The Machines== | |
| - | |||
| - | |||
| - | + | ===Lacto-detect=== | |
| - | |||
| - | + | This genetic machine within a population of bacteria will prime these bacteria to act as ‘Detectors’. It causes the production of a quorum signalling peptide in response to an input signal, which activates another machine, Lacto-report. | |
| - | + | [[Image:Wits_Machine_1_wiki.jpg]] | |
| - | + | '''Figure 1. The Lacto-sense construct''' | |
| - | + | ||
| - | + | ||
| - | ''' | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | So how did we end up selecting the final parts of our Machines? Each one was chosen after much consideration and scouring of the literature to select the most suitable biological system. For a design rationale of why we selected the parts that we did to create this machine, click [https://2010.igem.org/Team:WITS-South_Africa/machine1_design here] | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | All machines were constructed via sequential cloning steps in ''E.coli'' using the Standard Assembly method. | ||
| - | |||
| + | ===Lacto-report=== | ||
| - | + | This genetic machine is present in a second bacterial population, mixed in with the first. The quorum peptide produced when Lacto-sense is activated will be imported into the bacterial cells containing this machine and induce expression of this construct. This will have several effects: | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | * Production of more quorum peptide, to activate neighbouring cells and produce a positive feedback loop | ||
| + | * Production of a chromogenic reporter compound which is visible to the naked eye | ||
| + | * Production of a negative regulator which will block the binding of the quorum peptide and create a negative feedback loop, resetting the system to a negative state | ||
| - | |||
| + | [[Image:Wits_Machine_2.jpg]] | ||
| - | + | '''Figure 2. The Lacto-report construct''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | - | + | For a design rationale of why we selected the parts that we did to create this machine, click [https://2010.igem.org/Team:WITS-South_Africa/machine2_design here] |
| + | We were not able to finish constructing Lacto-report in its entirety, due to time constraints and issues with the E.chromi Biobrick parts. (For more details, click [https://2010.igem.org/Team:WITS-South_Africa/e-chromi here].) | ||
| - | + | For testing and characterising the behaviour of the quorum sensing mechanism in a model Gram-positive bacillus, we used an intermediate machine construct we dubbed Lacto-test. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | - | + | ===Lacto-test=== |
| - | + | This construct is a simplified of version of Lacto-report. When transformed into a Gram-positive bacteria, the presence of the PlcR promoter will allow us to determine if the PlcR-PapR quorum peptide can be correctly processed, imported into the cell and whether or not it activates the promoter. This can be determined by visualising expression of mCherry in a mixed culture of Lacto-detect and Lacto-report, after the addition of an inducer for Lacto-detect. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | - | + | [[Image:Wits_Lacto-test.jpg]] |
| - | + | '''Figure 3. The Lacto-test construct''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | --- | + | Lacto-test was cloned into the Gram-positive/E.coli shuttle vector pNDW5 and electroporated into a model Gram-positive bacillus for testing via fluorescent microscopy. |
| - | |||
| - | |||
| - | |||
| - | |||
| + | ==Machine Testing== | ||
| + | <br /> | ||
| - | + | All of our constructs contained different fluorescent protein markers. We added these in order to gain some preliminary data on the behaviour of our machines using fluorescent microscopy. Images were obtained by laser excitation of the fluorescent proteins at their excitation maximum wavelengths and filtered through a yellow/green channel to observe Venus or a red channel to observe mCherry. | |
| - | |||
| - | |||
| - | |||
| - | |||
| + | A culture of 2ml of ''E. coli'' containing Lacto-detect was grown overnight at 37°C and then transferred to 25 ml of ampicillin LB broth. This culture was then grown at 37°C in a shaking incubator for 2.5 hours so as to ensure that the bacteria were in their exponential growth phase. A baseline reading (Fig 4) was taken at this point (Tile A), to determine the presence of non-specific promoter activation. 10% 1mM IPTG was then added and the culture returned to the shaking incubator. An aliquot was then imaged after 30min (Tile B), and again after 1 hour (Tile C). All aliquots used in imaging were 100 ul. | ||
| + | As is seen in the baseline image (Tile A), the degree of fluorescent activation is minimal before the addition of IPTG. Tile B shows the fluorescent activation after 30min incubation with IPTG - there is very little visible fluorescent activation. After an hour of incubation (Tile C) there is a marked increase in the degree of fluorescence, thus providing an indication that IPTG has a positive effect on the Lac/Ara-1 promoter, activating gene expression. <br /> <br /> | ||
| + | [[Image:Montage_of_M1I_d.jpg|850px]] <br /> | ||
| + | '''Figure 4. Time course fluorescent microscopy of Lacto-detect before and after induction with IPTG''' <br /> <br /> <br /> | ||
| + | Once the induction of Lacto-detect had reached an appreciable level, it was added to an aliquot of ''B. subtilis'' containing the Lacto-test construct and imaged(Fig 5). Baseline images of the combined Lacto-detect and Lacto-test populations are shown. Tile A shows the Lacto-detect construct which has been activated (Imaged on a GFP filter which will only detect yellow/green fluorescence) and Tile B visualised red fluorescence which is indicative of activated mCherry in Lacto-report. Tile C is the same images overlaid to show a combined fluorescence profile of the sample. The degree of activation (Tile A) of Lacto-detect is appreciable due to the previous activation with IPTG, while the observed activation of Lacto-report (Tile B) shows no significant observable induction. <br /> <br /> | ||
| + | [[Image:Montage_of_Mixed.jpg|850px]] <br /> | ||
| + | '''Figure 5. Imaging of a mixed population of induced Lacto-detect with Lacto-test, immediately after the cultures were combined''' <br /> <br /> <br /> | ||
| + | Once the induced aliquot of Lacto-detect was added to Lacto-test, the combined aliquot was imaged at 30min intervals for 6 hours so as to observe the possible activation of Lacto-test by Lacto-detect (Figure 6). Figure 7 shows the imaged aliquot from Figure 6 with the Venus and mCherry fluorescent channels separated and presented at each epoch as alphabetical sets - A&B, C&D etc. This is done as to fully appreciate the direct induction of Lacto-test by Lacto-detect. Taking particular note of tile G of Figure 6 and the corresponding tiles M&N of Figure 7 (after 3.5h), there is marked increase in both Venus and mCherry fluorescence. This is indicative of the active communication between Lacto-detect and Lacto-test, thus illustrating the efficacy of the PlcR-PapR quorum network. <br /> <br /> | ||
| + | [[Image:Montage_of_SuperMontage.jpg|850px]] <br /> '''Figure 6. Combined Lacto-test and Lacto-report populations imaged over time to demonstrate induction''' <br /> <br /> | ||
| + | [[Image:Montage_of_Combo.jpg|850px]] <br /> '''Figure 7. Combined Lacto-test and Lacto-report populations imaged over time to demonstrate induction, showing the seperate fluorescent channels''' | ||
| + | ==Conclusions== | ||
| - | |||
| - | + | This data is promising, indicating that a quorum sensing mechanism from one Gram-positive organism such as ''Bacillus thuriengiensis'' can potentially be imported into another, such as ''Bacillus subtilis'', and successfully allows for co-ordinated control of gene expression. | |
| - | + | ||
| - | |||
| + | It also indicates that it is possible to obtain functional quorum peptides from a Gram-negative such as ''E.coli'', which would ordinarily not produce these molecules. | ||
| - | |||
| - | + | The synthetic PlcR-PapR fusion peptide we created appears to be fully functional in it's ability to bind to the PlcR promoter and activate gene expression. | |
| - | |||
| - | + | Both the Lac1/AraC promoter and the PlcR promoter appear to be fairly tightly regulated, with very little baseline expression, making them promising candidates as inducible promoters. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | + | '''The PlcR promoter and PlcR-PapR protein system is thus a useful inducible gene expression system for Gram-positive bacteria and can be used in the most commonly used Gram-positive chassis, ''Bacillus subtilis''.''' | |
Latest revision as of 21:01, 27 October 2010

Contents |
The Machines
Lacto-detect
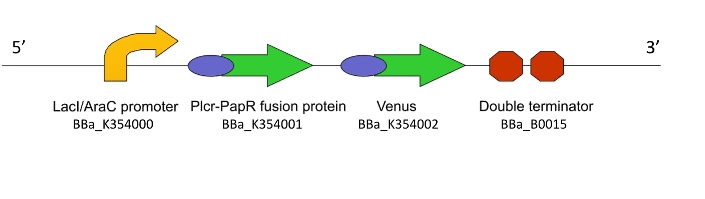
This genetic machine within a population of bacteria will prime these bacteria to act as ‘Detectors’. It causes the production of a quorum signalling peptide in response to an input signal, which activates another machine, Lacto-report.
Figure 1. The Lacto-sense construct
So how did we end up selecting the final parts of our Machines? Each one was chosen after much consideration and scouring of the literature to select the most suitable biological system. For a design rationale of why we selected the parts that we did to create this machine, click here
All machines were constructed via sequential cloning steps in E.coli using the Standard Assembly method.
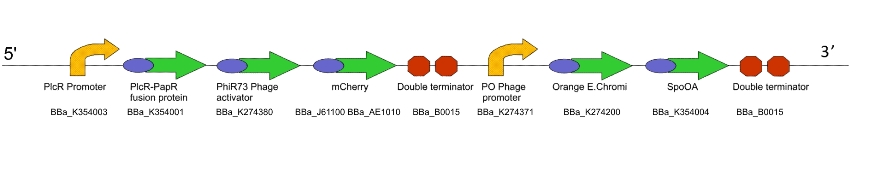
Lacto-report
This genetic machine is present in a second bacterial population, mixed in with the first. The quorum peptide produced when Lacto-sense is activated will be imported into the bacterial cells containing this machine and induce expression of this construct. This will have several effects:
- Production of more quorum peptide, to activate neighbouring cells and produce a positive feedback loop
- Production of a chromogenic reporter compound which is visible to the naked eye
- Production of a negative regulator which will block the binding of the quorum peptide and create a negative feedback loop, resetting the system to a negative state
Figure 2. The Lacto-report construct
For a design rationale of why we selected the parts that we did to create this machine, click here
We were not able to finish constructing Lacto-report in its entirety, due to time constraints and issues with the E.chromi Biobrick parts. (For more details, click here.)
For testing and characterising the behaviour of the quorum sensing mechanism in a model Gram-positive bacillus, we used an intermediate machine construct we dubbed Lacto-test.
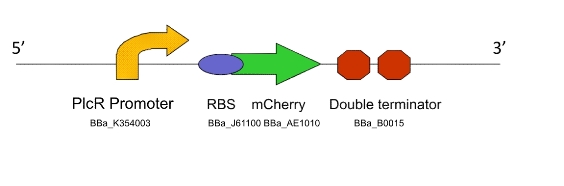
Lacto-test
This construct is a simplified of version of Lacto-report. When transformed into a Gram-positive bacteria, the presence of the PlcR promoter will allow us to determine if the PlcR-PapR quorum peptide can be correctly processed, imported into the cell and whether or not it activates the promoter. This can be determined by visualising expression of mCherry in a mixed culture of Lacto-detect and Lacto-report, after the addition of an inducer for Lacto-detect.
Figure 3. The Lacto-test construct
Lacto-test was cloned into the Gram-positive/E.coli shuttle vector pNDW5 and electroporated into a model Gram-positive bacillus for testing via fluorescent microscopy.
Machine Testing
All of our constructs contained different fluorescent protein markers. We added these in order to gain some preliminary data on the behaviour of our machines using fluorescent microscopy. Images were obtained by laser excitation of the fluorescent proteins at their excitation maximum wavelengths and filtered through a yellow/green channel to observe Venus or a red channel to observe mCherry.
A culture of 2ml of E. coli containing Lacto-detect was grown overnight at 37°C and then transferred to 25 ml of ampicillin LB broth. This culture was then grown at 37°C in a shaking incubator for 2.5 hours so as to ensure that the bacteria were in their exponential growth phase. A baseline reading (Fig 4) was taken at this point (Tile A), to determine the presence of non-specific promoter activation. 10% 1mM IPTG was then added and the culture returned to the shaking incubator. An aliquot was then imaged after 30min (Tile B), and again after 1 hour (Tile C). All aliquots used in imaging were 100 ul.
As is seen in the baseline image (Tile A), the degree of fluorescent activation is minimal before the addition of IPTG. Tile B shows the fluorescent activation after 30min incubation with IPTG - there is very little visible fluorescent activation. After an hour of incubation (Tile C) there is a marked increase in the degree of fluorescence, thus providing an indication that IPTG has a positive effect on the Lac/Ara-1 promoter, activating gene expression.
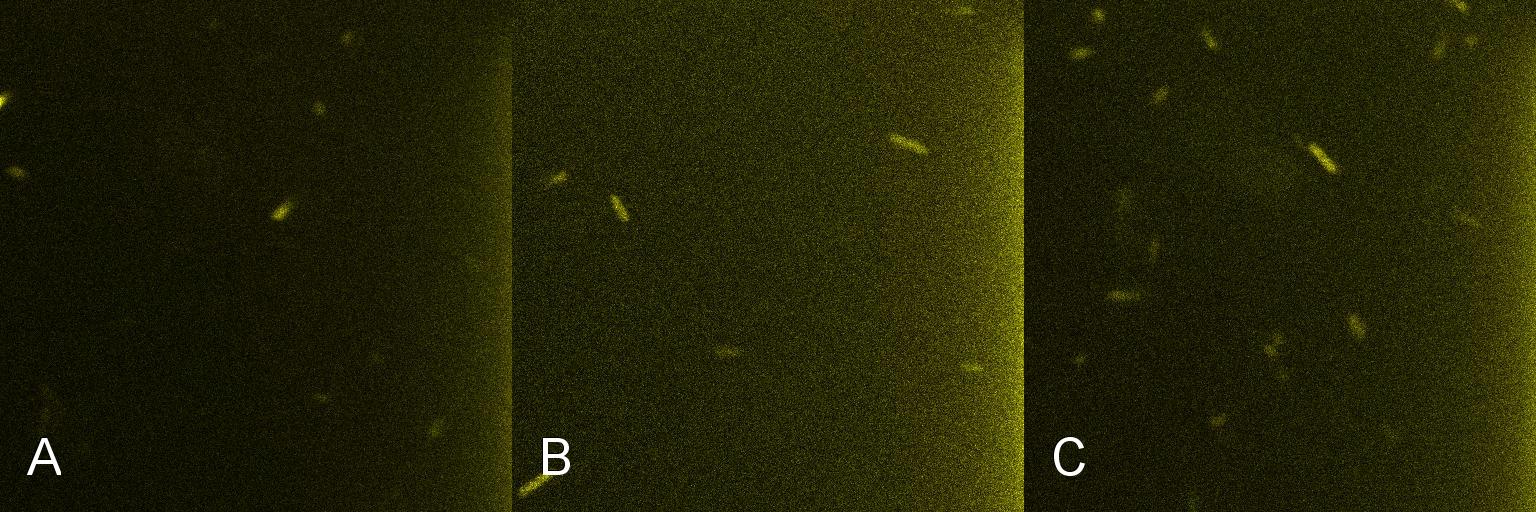
Figure 4. Time course fluorescent microscopy of Lacto-detect before and after induction with IPTG
Once the induction of Lacto-detect had reached an appreciable level, it was added to an aliquot of B. subtilis containing the Lacto-test construct and imaged(Fig 5). Baseline images of the combined Lacto-detect and Lacto-test populations are shown. Tile A shows the Lacto-detect construct which has been activated (Imaged on a GFP filter which will only detect yellow/green fluorescence) and Tile B visualised red fluorescence which is indicative of activated mCherry in Lacto-report. Tile C is the same images overlaid to show a combined fluorescence profile of the sample. The degree of activation (Tile A) of Lacto-detect is appreciable due to the previous activation with IPTG, while the observed activation of Lacto-report (Tile B) shows no significant observable induction.
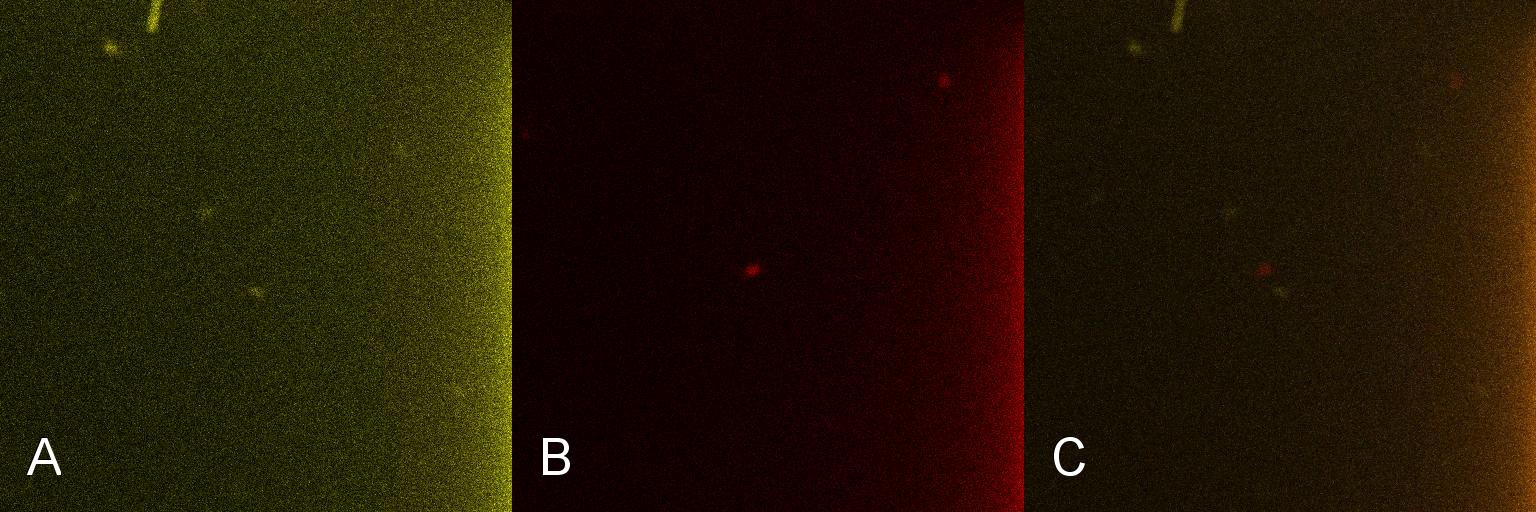
Figure 5. Imaging of a mixed population of induced Lacto-detect with Lacto-test, immediately after the cultures were combined
Once the induced aliquot of Lacto-detect was added to Lacto-test, the combined aliquot was imaged at 30min intervals for 6 hours so as to observe the possible activation of Lacto-test by Lacto-detect (Figure 6). Figure 7 shows the imaged aliquot from Figure 6 with the Venus and mCherry fluorescent channels separated and presented at each epoch as alphabetical sets - A&B, C&D etc. This is done as to fully appreciate the direct induction of Lacto-test by Lacto-detect. Taking particular note of tile G of Figure 6 and the corresponding tiles M&N of Figure 7 (after 3.5h), there is marked increase in both Venus and mCherry fluorescence. This is indicative of the active communication between Lacto-detect and Lacto-test, thus illustrating the efficacy of the PlcR-PapR quorum network.
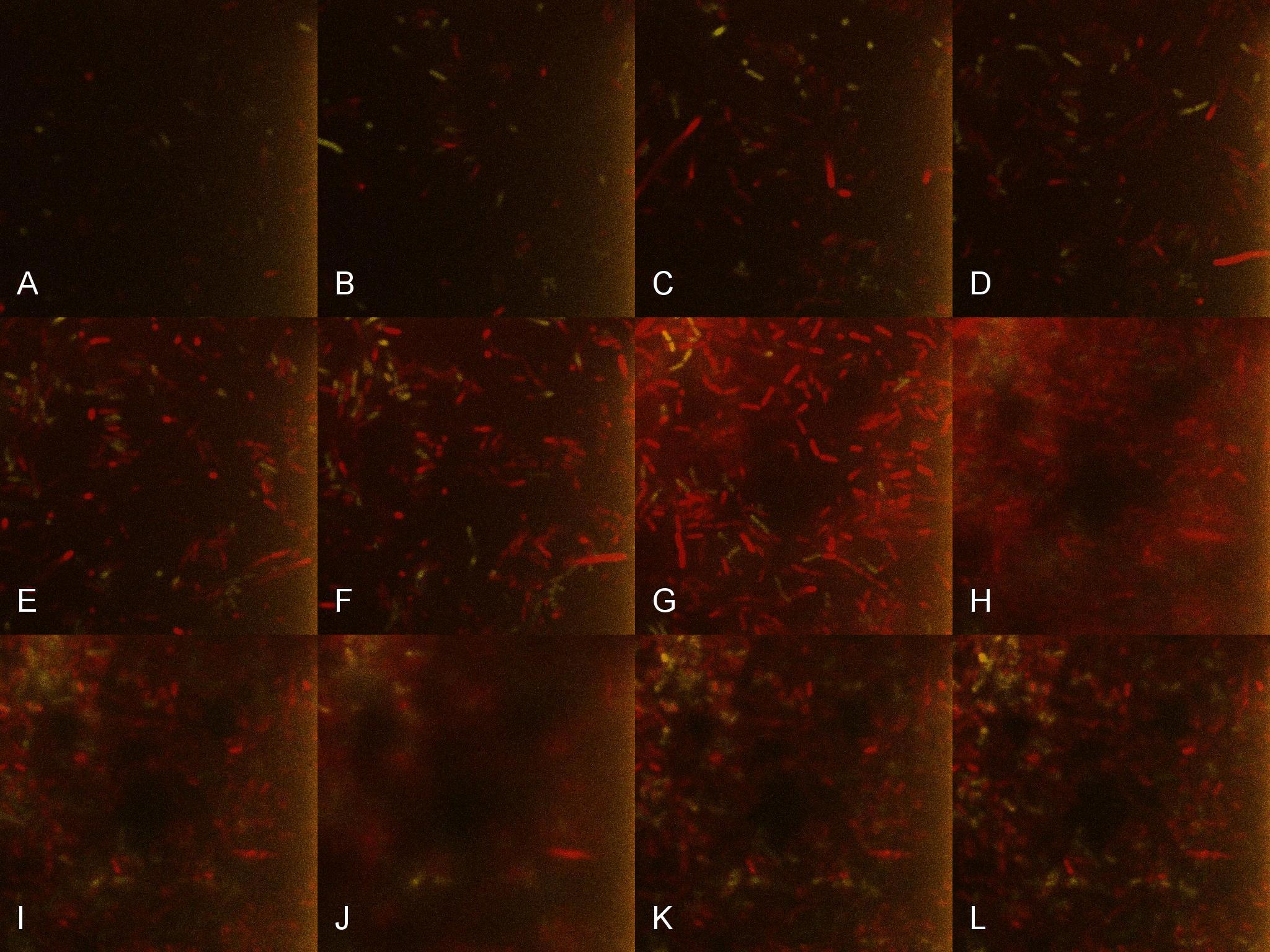
Figure 6. Combined Lacto-test and Lacto-report populations imaged over time to demonstrate induction
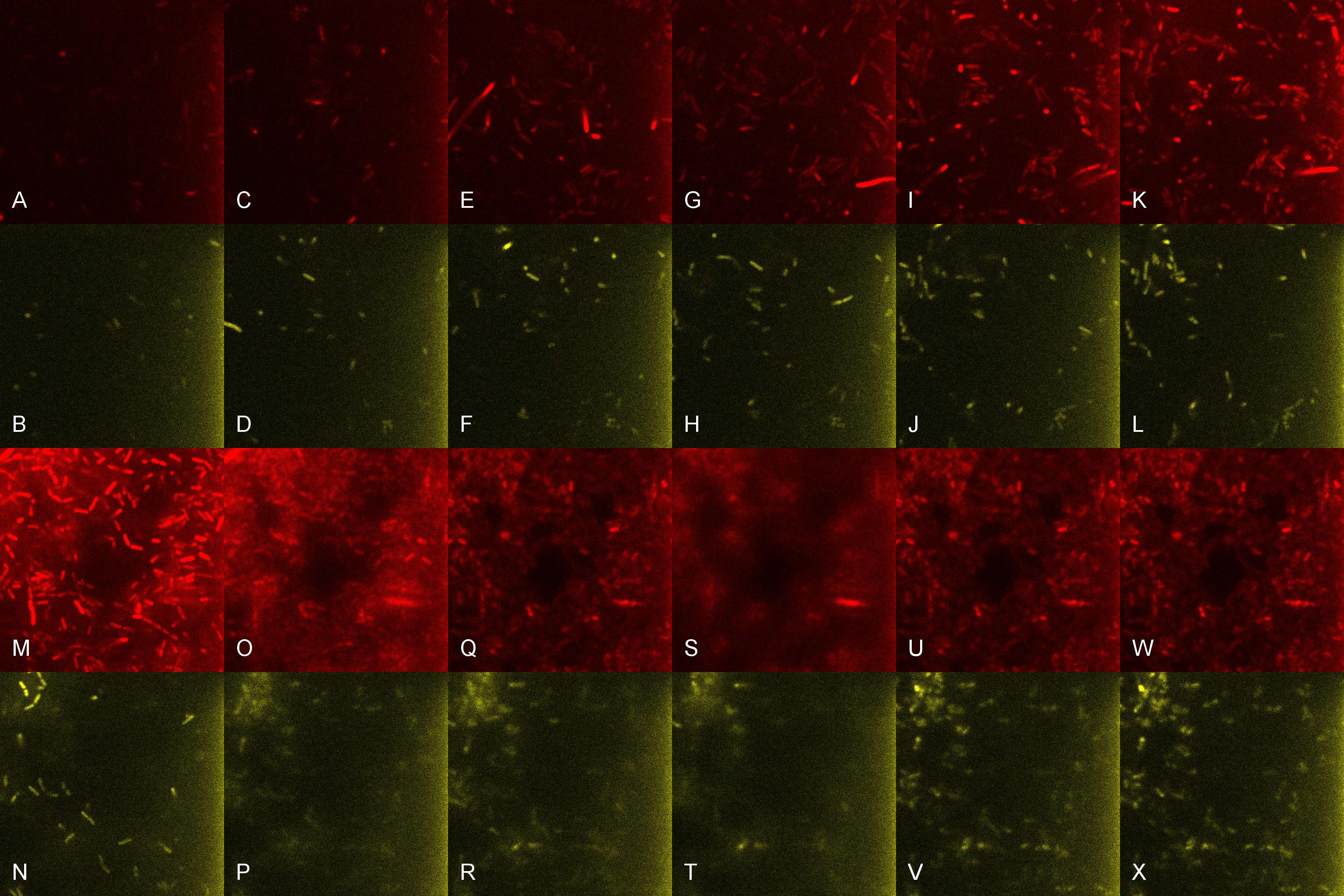
Figure 7. Combined Lacto-test and Lacto-report populations imaged over time to demonstrate induction, showing the seperate fluorescent channels
Conclusions
This data is promising, indicating that a quorum sensing mechanism from one Gram-positive organism such as Bacillus thuriengiensis can potentially be imported into another, such as Bacillus subtilis, and successfully allows for co-ordinated control of gene expression.
It also indicates that it is possible to obtain functional quorum peptides from a Gram-negative such as E.coli, which would ordinarily not produce these molecules.
The synthetic PlcR-PapR fusion peptide we created appears to be fully functional in it's ability to bind to the PlcR promoter and activate gene expression.
Both the Lac1/AraC promoter and the PlcR promoter appear to be fairly tightly regulated, with very little baseline expression, making them promising candidates as inducible promoters.
 "
"