Team:WITS-South Africa/Gram positives
From 2010.igem.org
(→Methods) |
(→Introduction) |
||
| (15 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{{Template:WITS-South_Africa_Main_Menu_0910}} | {{Template:WITS-South_Africa_Main_Menu_0910}} | ||
| + | <div style="padding:40px;"> | ||
| - | <div class=" | + | <div class="heading"> |
| - | Gram | + | Gram-positive bacteria |
| + | ---- | ||
</div> | </div> | ||
| + | |||
<div style="padding:10px;"> | <div style="padding:10px;"> | ||
<div style="width:850px;text-align:justify;float:middle;"> | <div style="width:850px;text-align:justify;float:middle;"> | ||
| Line 15: | Line 18: | ||
We have been working on optimising a protocol for electro-transformations on several gram positive bacteria that can function as the chassis for our machine. | We have been working on optimising a protocol for electro-transformations on several gram positive bacteria that can function as the chassis for our machine. | ||
| - | Lange has been performing several test experiments by electroporating several potential gram positive bacteria (i.e. ''Bacillus subtilis''; ''Lactobacillus gasseri''and ''Staphylococcus aureus''). This is being done in preparation for electroporation of the machine constructs into either one of these gram positive bacteria; depending on the most efficient bacteria we have to work with. | + | Lange has been performing several test experiments by electroporating several potential gram positive bacteria (i.e. ''Bacillus subtilis''; ''Lactobacillus gasseri'' and ''Staphylococcus aureus''). This is being done in preparation for electroporation of the machine constructs into either one of these gram positive bacteria; depending on the most efficient bacteria we have to work with. |
We are also testing the ability of transforming the above mentioned bacteria with two different shuttle-vector plasmids; namely: | We are also testing the ability of transforming the above mentioned bacteria with two different shuttle-vector plasmids; namely: | ||
| - | + | *pGK12 | |
| - | + | *pNDW5 | |
Once we have established which plasmid is best to use for electroporation; the gene constructs will then be inserted into that plasmid and we will perform electroporations using that plasmid. Electro-transformation of the selected bacterium will be done with our machine constructs; and testing of transformants will be undertaken. Once this is achieved analysis of the machines’ functionality will be determined using fluorescence in situ hybridisation and other fluorescent techniques (i.e. fluorimetry). | Once we have established which plasmid is best to use for electroporation; the gene constructs will then be inserted into that plasmid and we will perform electroporations using that plasmid. Electro-transformation of the selected bacterium will be done with our machine constructs; and testing of transformants will be undertaken. Once this is achieved analysis of the machines’ functionality will be determined using fluorescence in situ hybridisation and other fluorescent techniques (i.e. fluorimetry). | ||
| Line 135: | Line 138: | ||
---- | ---- | ||
| - | == '''Test 2. Electroporation of pNDW5* and pGK12 into L. gasseri''' == | + | == '''Test 2. Electroporation of pNDW5* and pGK12 into'' L. gasseri''''' == |
| Line 149: | Line 152: | ||
- The concentrations used for the antibiotic plates were as follows according to the above protocol: | - The concentrations used for the antibiotic plates were as follows according to the above protocol: | ||
| + | |||
• Erythromycin (Ery) 1 µg/ml | • Erythromycin (Ery) 1 µg/ml | ||
| - | • Erythromycin 1 µg/ml and Chloroamphenicol 5 µg/ml | + | |
| + | • (Erythromycin 1 µg/ml and Chloroamphenicol 5 µg/ml) | ||
| + | |||
• Chloroamphenicol (Cm) 5 µg/ml | • Chloroamphenicol (Cm) 5 µg/ml | ||
- The media used, Electroporator parameters and incubation styles are different for ''L. gasseri'' | - The media used, Electroporator parameters and incubation styles are different for ''L. gasseri'' | ||
| + | |||
• Media = MRS for broth and agar; Sucrose Media | • Media = MRS for broth and agar; Sucrose Media | ||
| - | • Electroporation buffer = MRSSM (MRS in Sucrose | + | |
| + | • Electroporation buffer = MRSSM (MRS in Sucrose | ||
| + | |||
• Electroporator parameters: 1.5 kV; 800 Ω; 25µF | • Electroporator parameters: 1.5 kV; 800 Ω; 25µF | ||
| + | |||
• Growth = anaerobic chamber and not shaken | • Growth = anaerobic chamber and not shaken | ||
=== Results === | === Results === | ||
| - | The growth | + | The growth kinetics for ''L. gasseri'' are shown below; these illustrate the rate in which ''L. gasseri'' naturally grows in suitable growth media. the ''L. gasseri'' used for kinetics was untransformed: |
[[Image:L._gaseri_growth_curve.png]] | [[Image:L._gaseri_growth_curve.png]] | ||
| Line 180: | Line 190: | ||
It can be seen from Table 2 that the pGK12 Ery (1µg/ml) plate; and Ery 1µg/ml Cm 5µg/ml plate had colonies that were possible transformants growing on them and so plasmid preps were undertaken. | It can be seen from Table 2 that the pGK12 Ery (1µg/ml) plate; and Ery 1µg/ml Cm 5µg/ml plate had colonies that were possible transformants growing on them and so plasmid preps were undertaken. | ||
| - | The transformation efficiency could not be obtained for pGK12 because the original pGK12 DNA sample was either degraded or its concentration was very low. When the original pGK12 sample was transformed into ''E. coli''; colony numbers were low (2 colonies) whereas a control plasmid (pACYC184) gave a high number of colonies. | + | The transformation efficiency could not be obtained for pGK12 because the original pGK12 DNA sample was either degraded or its concentration was very low (>1ng/µl). |
| + | |||
| + | When the original pGK12 sample was transformed into ''E. coli''; colony numbers were low (2 colonies) whereas a control plasmid (pACYC184) gave a high number of colonies. | ||
pNDW5* shows 80 colonies on the erythromycin (1µg/ml) plate. This was obviously an error as pNDW5 does not confer resistance to erythromycin; therefore the colonies obtained shouldn’t have been observed here. | pNDW5* shows 80 colonies on the erythromycin (1µg/ml) plate. This was obviously an error as pNDW5 does not confer resistance to erythromycin; therefore the colonies obtained shouldn’t have been observed here. | ||
The transformation efficiency could not be measured here due to the inconsistency of the pNDW5* results. | The transformation efficiency could not be measured here due to the inconsistency of the pNDW5* results. | ||
| Line 187: | Line 199: | ||
---- | ---- | ||
| - | + | == '''Test 3. Electroporation of pNDW5* and pGK12 into'' B. subtilis'' and ''S. aureus''''' == | |
| - | == '''Test 3. Electroporation of pNDW5* and pGK12 into B. subtilis and S. aureus''' == | + | |
=== Methods === | === Methods === | ||
| Line 206: | Line 217: | ||
---- | ---- | ||
| - | + | == '''Test 4 Electroporation of machine cassettes in'' B. subtilis'''''== | |
| - | == '''Test 4 Electroporation of machine cassettes in B. subtilis'''== | + | |
=== Introduction === | === Introduction === | ||
| Line 218: | Line 228: | ||
=== Methods === | === Methods === | ||
| - | The | + | The protocol used for electroporating into ''B. subtilis'' was obtained from: |
| + | |||
| + | Stephenson, M. and Jarrett, P. (1991). Transformation of ''Bacillus subtilis'' by Electroporation. ''Biotechnology Techniques''. Vol '''5''' (1); 9-12 | ||
| + | |||
[[Image:Electroporation process.png]] | [[Image:Electroporation process.png]] | ||
| Line 224: | Line 237: | ||
'''Figure 1: Flow diagram representing the transfromation process of ''B. subtilis by Electroporation''''' | '''Figure 1: Flow diagram representing the transfromation process of ''B. subtilis by Electroporation''''' | ||
| - | ''B. subtilis'' strains were grown overnight in LB broth at 37°C with vigorous aeration. The changes to the Electroporation protocol included: | + | ''B. subtilis'' strains were grown overnight in LB broth at 37°C with vigorous aeration. The changes to the '''General''' Electroporation protocol included: |
1. SOC Broth= pulsed cells were diluted in 2ml of this broth(contains 2% tryptone, 0.5% yeast extract, 10mM NaCl, 2.5mM KCl, 10mM MgCl2, 10mM MgSO4, 20mM glucose) | 1. SOC Broth= pulsed cells were diluted in 2ml of this broth(contains 2% tryptone, 0.5% yeast extract, 10mM NaCl, 2.5mM KCl, 10mM MgCl2, 10mM MgSO4, 20mM glucose) | ||
| + | |||
2. 30% PEG6000 = Polyethyleneglycol (Electroporation medium) | 2. 30% PEG6000 = Polyethyleneglycol (Electroporation medium) | ||
| + | |||
3. De – ionised water for the washes. | 3. De – ionised water for the washes. | ||
| Line 234: | Line 249: | ||
The following constructs were electroporated. | The following constructs were electroporated. | ||
| - | - Machine Intermediate 1 (LacI/ AraC promoter – mCherry – tt) | + | - Machine Intermediate 1 (LacI/ AraC promoter – mCherry – tt) in pNDW5 |
| - | - Machine Intermediate 2 (PlcR promoter – mCherry – tt) | + | - Machine Intermediate 2 (PlcR promoter – mCherry – tt) in pNDW5 |
- pNDW5* vector (no “machine” genetic information; control) | - pNDW5* vector (no “machine” genetic information; control) | ||
| Line 242: | Line 257: | ||
- Lactodetect ( LacI/ AraC promoter – PlcR –PapR – Venus- tt) in pNDW5 | - Lactodetect ( LacI/ AraC promoter – PlcR –PapR – Venus- tt) in pNDW5 | ||
| - | - Lactoreport (PlcR promoter – PlcR-PapR- mCherry – PO Phage activator – tt – PO Phage promoter – E. chromi – SpooA – tt) | + | - Lactoreport (PlcR promoter – PlcR-PapR- mCherry – PO Phage activator – tt – PO Phage promoter – E. chromi – SpooA – tt) in pNDW5 |
| + | |||
==== Mass calculation for machine 1 and 2 intermediates ==== | ==== Mass calculation for machine 1 and 2 intermediates ==== | ||
| - | Machine 1 intermediate DNA conc. = 52.2ng/µl; Machine 2 intermediate DNA conc. = 48.3ng/ml | + | |
| - | The volume used for electroporation was 5µl | + | |
| + | Machine 1 intermediate DNA conc. = '''52.2ng/µl'''; Machine 2 intermediate DNA conc. = '''48.3ng/ml''' | ||
| + | The volume used for electroporation was '''5µl''' | ||
Hence: C = M / V whereby C is the concentration, M is mass and V is volume | Hence: C = M / V whereby C is the concentration, M is mass and V is volume | ||
| - | Mass of Machine 1 intermediate =C x V = 52.2ng/µl x 5µl = 261ng | + | Mass of Machine 1 intermediate =C x V = 52.2ng/µl x 5µl = '''261ng''' |
| + | |||
| + | Mass of Machine 2 intermediate = C x V = 48.3ng/µl x 5µl = '''241.5ng''' | ||
| + | |||
| + | |||
| + | ==== Electroporation Efficiencies for Machine intermediate 1 and 2 in ''B. subtilis'' ==== | ||
| - | |||
| - | |||
Colonies from electroporation plates were counted and yielded the following: | Colonies from electroporation plates were counted and yielded the following: | ||
| - | Machine | + | Machine intermediate 1 = '''1300 transformants''' |
| - | Machine | + | Machine intermediate 1 electroporation efficiency = 1300 colonies/261ng DNA = 1300/0.261µg DNA = '''4980 colonies/ µg DNA''' |
| - | Machine | + | Machine intermediate 2 = '''1400 transformants''' |
| - | Machine | + | Machine intermediate 2 electroporation efficiency = 1400 colonies/241.5ng DNA = 1400/0.2415µg DNA = '''5797 colonies/ µg DNA''' |
| - | pNDW5* = 0 transformants (No DNA control) | + | pNDW5* = '''0 transformants''' (No DNA control) |
The pNDW5* vector had no transformants as expected | The pNDW5* vector had no transformants as expected | ||
Electroporation efficiencies were not calculated for Lactodetect and Lactoreport | Electroporation efficiencies were not calculated for Lactodetect and Lactoreport | ||
Latest revision as of 21:15, 27 October 2010

Gram-positive bacteria
Contents
|
Introduction
We have been working on optimising a protocol for electro-transformations on several gram positive bacteria that can function as the chassis for our machine. Lange has been performing several test experiments by electroporating several potential gram positive bacteria (i.e. Bacillus subtilis; Lactobacillus gasseri and Staphylococcus aureus). This is being done in preparation for electroporation of the machine constructs into either one of these gram positive bacteria; depending on the most efficient bacteria we have to work with.
We are also testing the ability of transforming the above mentioned bacteria with two different shuttle-vector plasmids; namely:
- pGK12
- pNDW5
Once we have established which plasmid is best to use for electroporation; the gene constructs will then be inserted into that plasmid and we will perform electroporations using that plasmid. Electro-transformation of the selected bacterium will be done with our machine constructs; and testing of transformants will be undertaken. Once this is achieved analysis of the machines’ functionality will be determined using fluorescence in situ hybridisation and other fluorescent techniques (i.e. fluorimetry).
We plan to achieve a high efficiency of gram positive transformation by optimizing relevant protocols for electroporation and fluorescence analysis.
Why Electroporation?
Electro-transformations are mostly used to transform bacteria using large amounts of plasmid DNA; this method involves the use of an electric pulse that creates temporary pores in the cell wall of bacteria allowing the bacteria to take up the plasmid. Initially this plasmid needs to be along the surface of the cell wall for instant incorporation of the plasmid into the bacterial cytoplasm when an electric pulse in administered. Protocols for electro-transforming different kinds of gram positive bacteria vary depending on the strain, or family of bacteria (i.e. Lactobacillus species have been shown to require different media for Electroporation).
Electro-transformation of potential Gram positive bacteria
The Gram positive bacteria that are being tested are:
1. Bacillus subtilis
2. Lactobacillus gasseri
3. Staphyloccus aureus
4. An unknown Lactobacillus strain
These bacteria have been selected because we would ideally like the chassis for our whole cell biosensor to be a gram positive bacterium. This being said; women have commensal lactobacillus gasseri bacteria within their vaginal mucosa. This bacterium provides the ideal target for genetically engineering L. gasseri to act as a proxy for infection and subsequently for amplifying the quorum signal and propagating the signal for infection across the population of bacterial cells.
B. subtilis, S aureus and the LAB sp. Are also being used considering their gram positive properties. They are being tested to determine whether or not they could substitute L. gasseri as a chassis for our machine; only if Electroporation of L. gasseri fails.
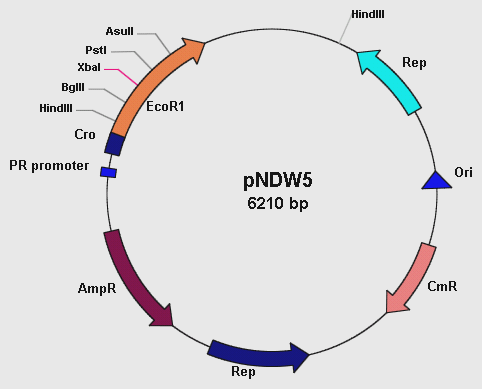
pNDW5 Plasmid
Figure 1: showing the pNDW5 plasmid map.
The pNDW5 plasmid is derived from a combination of two bacterial plasmids hence a shuttle vector; whereby one is gram positive and the other gram negative (S. aureus and E. coli respectively). The S. aureus plasmid pC194 and E. coli plasmid pEcoR251 were combined to form the pNDW5 plasmid. This pNDW5 plasmid has a separate pair of Ori and Rep for gram positive and gram negative bacteria. This plasmid contains ampicillin and chloroamphenicol resistance marker genes; therefore ampicillin and chloroamphenicol resistant transformants with this plasmid can be selected. The pNDW5 plasmid has an EcoRI suicide gene instead of a multiple cloning region; inside the gene are 4 unique restriction sites which allows for the positive selection of transformants with inserts. The plasmid has a PR promoter (phage λ promoter) whose original function is to drive the transcription of the Cro gene which allows for induction of phage from its lysogenic state in the E. coli cell. A large portion of this Cro gene has been removed which defers its functionality allowing the use of the PR promoter to drive expression of EcoRI without expressing the Cro gene. In pNDW5, a repressor can be used to stop transcription and expression of EcoRI suicide gene. The EcoRI suicide gene automatically kills the cell if no genes have been cloned. For testing the vector a mutant of this pNDW5 plasmid was used (denoted pNDW5*); which allowed testing for electro-transformation qualities in various potential gram positive bacteria (B. subtilis, L. gasseri, S. aureus and a LAB sp).
pGK12 plasmid
The pGK12 plasmid is also a shuttle vector; it can be propagated in both Gram-positive and Gram-negative bacteria and it carries erythromycin and chloroamphenicol resistance markers. This plasmid has 3 restriction sites; HpaII, NdeI and BpmI. A diagram for this plasmid has not been put up due to the poor quality of pGK12 diagrams that have been made available to us.
What has been happening in the lab
Electroporation of potential Gram positive bacteria
The protocols for electroporating all gram positive bacteria used in our project except L. gasseri can be obtained from http://openwetware.org/wiki/Main_Page
General Electroporation protocol
• Preparing electro-competent cells
• Overnight culturing in Luria Broth (LB)*
• Inoculate x100 dilution of culture into LB with 1.9% glycine
• Incubate at 37°C
• Measure 0.D. 600 (range 0.7)
• Harvest cells (centrifuge)
• Wash cells (x2) in Electroporation buffer
• Harvest cells and suspend in 1/100 volume of Electroporation buffer
• 50µl transformation
• Electroporator parameters vary depending on strain
• Inoculate in LB and incubate for 3hours at 37°C on a shaker
• Plate in Luria agar and appropriate antibiotic plate
(Note: LB* = media may vary depending on bacterial strain or species)
The tests that I am currently performing are as follows:
Test 1. Electroporation of pNDW5* into a Lactobacillus sp.
Methods
The above protocol was administered for electroporating the unknown LAB sp. There were no changes made. Electroporator parameters: 1.5 kV; 200 Ω; 25µF
Results
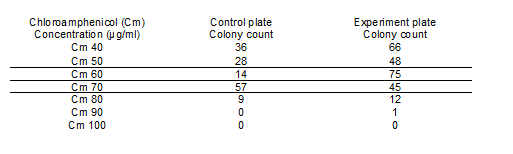
Upon completing Electroporation the following assessments were made: The control and experimental plates had an equal number of colonies when chloroamphenicol (Cm) was used as a resistance marker at 20µg/ml. Hence secondary testing for the appropriate chloroamphenicol concentration was done.
Table 1: LAB colonies in the control and experimental plates at increasing chloroamphenicol concentration. (Note: control plates are the LAB only; experiment plate has electroporated LAB). From these colonies further tests on transformants were done. Chloroamphenicol (Cm)
At Cm concentrations of 60 µg/ml the ratio of experimental colonies to control colonies was approximately 5:1. To verify whether these colonies were real transformants plasmid extractions were attempted from these cells. However no plasmid could be detected. These results also suggest that this LAB strain has inherent resistance to chloroamphenicol therefore it cannot be used to determine whether pNDW5 would transform the LAB sp.
Transformation efficiency
In order to determine the success of transformation, the concentration of DNA used for the transformation would be determined. There are two methods to do this.
1. The first is to analyze band intensities after gel electrophoresis in comparison to a control band whose DNA concentration is already known.
2. The alternative method is to measure DNA concentration with a spectrophotometer at 260nm. Transformation efficiency would be expressed as the number of transformants per µg of DNA.
Test 2. Electroporation of pNDW5* and pGK12 into L. gasseri
Methods
- A standard curve for analysing the growth characteristics of L. gasseri was undertaken
- The protocol for electroporating L. gasseri was obtained from:
Nickoloff, J.A. (2007). Electroporation Protocols for Microorganisms: chapter 20- Transformation of Lactobacillus by Electroporation. Methods in Molecular Biology 47: 201- 208
- The appropriate media was made; preparation of competent cells and Electroporation was done using the above mentioned protocol with minor adjustments. The most common marker genes applicable to LAB are genes causing resistance to erythromycin (Ery) and chloroamphenicol (Cm).
- The concentrations used for the antibiotic plates were as follows according to the above protocol:
• Erythromycin (Ery) 1 µg/ml
• (Erythromycin 1 µg/ml and Chloroamphenicol 5 µg/ml)
• Chloroamphenicol (Cm) 5 µg/ml
- The media used, Electroporator parameters and incubation styles are different for L. gasseri
• Media = MRS for broth and agar; Sucrose Media
• Electroporation buffer = MRSSM (MRS in Sucrose
• Electroporator parameters: 1.5 kV; 800 Ω; 25µF
• Growth = anaerobic chamber and not shaken
Results
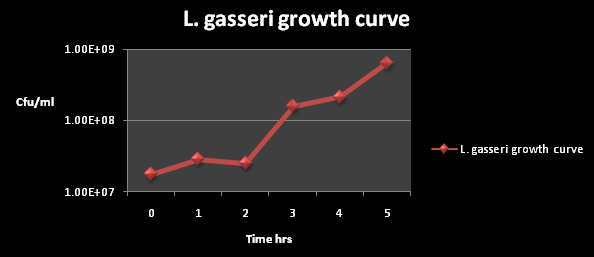
The growth kinetics for L. gasseri are shown below; these illustrate the rate in which L. gasseri naturally grows in suitable growth media. the L. gasseri used for kinetics was untransformed:
Figure 1: The cfu/ml of L. gasseri over a time period of 5 hours. The graph suggests an exponential increase in the colony forming units/ml after 2 hours of incubation.
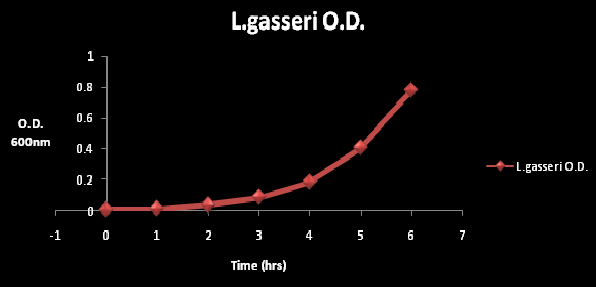
Figure 2: The O.D at wavelength of 600nm at time period of 6 hours. The O.D. was measured every hour. The gradual increase in O.D. is reflective of cell growth.
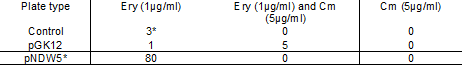
Table 2: Shows the Electroporation colony numbers for the pGK12, pNDW5 plasmid and the control plate at the different antibiotic concentrations
Note: the 3 colonies in the Ery 1µg/ml control plate were all contaminants and were not L. gasseri.
It can be seen from Table 2 that the pGK12 Ery (1µg/ml) plate; and Ery 1µg/ml Cm 5µg/ml plate had colonies that were possible transformants growing on them and so plasmid preps were undertaken.
The transformation efficiency could not be obtained for pGK12 because the original pGK12 DNA sample was either degraded or its concentration was very low (>1ng/µl).
When the original pGK12 sample was transformed into E. coli; colony numbers were low (2 colonies) whereas a control plasmid (pACYC184) gave a high number of colonies. pNDW5* shows 80 colonies on the erythromycin (1µg/ml) plate. This was obviously an error as pNDW5 does not confer resistance to erythromycin; therefore the colonies obtained shouldn’t have been observed here. The transformation efficiency could not be measured here due to the inconsistency of the pNDW5* results.
Test 3. Electroporation of pNDW5* and pGK12 into B. subtilis and S. aureus
Methods
The general protocol obtained from http://openwetware.org/wiki/Main_Page was used to electroporate pNDW5* and pGK12 into B. subtilis and S. aureus.
A few changes were made to the protocol: • media used = LB • Electroporation buffer = SHMG (sucrose, HEPES, magnesium chloride and glycerol) buffer • Incubation period = overnight for B. subtilis
Results
Electroporation of S. aureus has been successful whereby optimization is being done for B. subtilis. Upon further optimization test 4 was conducted for transformation of B. subtilis by electroporation. there was no further experimental work done on S. aureus as B. subtilis was chosen an alternative model organism for L. gasseri. pNDW5 was futher electroporated in Test 4. No further testing was done in S. aureus as B. subtilis was prefered as an alternative chassis for the Lactoguard machine.
Test 4 Electroporation of machine cassettes in B. subtilis
Introduction
Bacillus subtilis is a gram positive soil bacterium from the genus bacillus. It has proven highly useful to genetic manipulation, and has therefore become widely adopted as a model organism for laboratory studies. It is also heavily flagellated, which gives B. subtilis the ability to move quite quickly. In terms of popularity as a laboratory model organism this bacterium is often used as the Gram-positive equivalent of Escherichia coli, an extensively studied Gram-negative bacterium.
It has proved useful in electroporating our machine constructs and has yielded good electroporation efficiencies for Machine Intermediates 1 and 2. Lactodetect and Lactoreport have also been successfully electro-transformed into B. subtilis as numerous transformants were present from colony plate counts however no empirical data has been calculated for these constructs. It is no surprise therefore that we have chosen to work with this bacterium as a model organism as an alternative to L. gasseri as a chassis for our Lactoguard Machine.
Methods
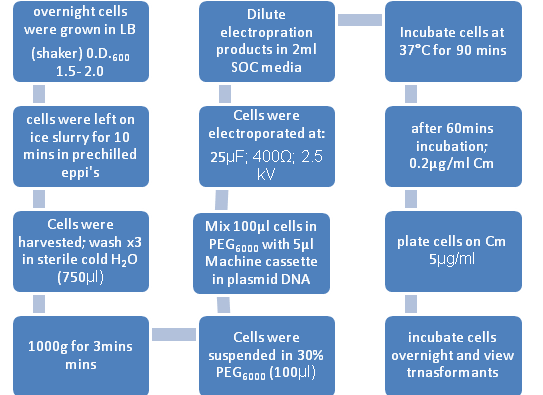
The protocol used for electroporating into B. subtilis was obtained from:
Stephenson, M. and Jarrett, P. (1991). Transformation of Bacillus subtilis by Electroporation. Biotechnology Techniques. Vol 5 (1); 9-12
Figure 1: Flow diagram representing the transfromation process of B. subtilis by Electroporation
B. subtilis strains were grown overnight in LB broth at 37°C with vigorous aeration. The changes to the General Electroporation protocol included:
1. SOC Broth= pulsed cells were diluted in 2ml of this broth(contains 2% tryptone, 0.5% yeast extract, 10mM NaCl, 2.5mM KCl, 10mM MgCl2, 10mM MgSO4, 20mM glucose)
2. 30% PEG6000 = Polyethyleneglycol (Electroporation medium)
3. De – ionised water for the washes.
Results
The following constructs were electroporated.
- Machine Intermediate 1 (LacI/ AraC promoter – mCherry – tt) in pNDW5
- Machine Intermediate 2 (PlcR promoter – mCherry – tt) in pNDW5
- pNDW5* vector (no “machine” genetic information; control)
- Lactodetect ( LacI/ AraC promoter – PlcR –PapR – Venus- tt) in pNDW5
- Lactoreport (PlcR promoter – PlcR-PapR- mCherry – PO Phage activator – tt – PO Phage promoter – E. chromi – SpooA – tt) in pNDW5
Mass calculation for machine 1 and 2 intermediates
Machine 1 intermediate DNA conc. = 52.2ng/µl; Machine 2 intermediate DNA conc. = 48.3ng/ml The volume used for electroporation was 5µl
Hence: C = M / V whereby C is the concentration, M is mass and V is volume
Mass of Machine 1 intermediate =C x V = 52.2ng/µl x 5µl = 261ng
Mass of Machine 2 intermediate = C x V = 48.3ng/µl x 5µl = 241.5ng
Electroporation Efficiencies for Machine intermediate 1 and 2 in B. subtilis
Colonies from electroporation plates were counted and yielded the following:
Machine intermediate 1 = 1300 transformants
Machine intermediate 1 electroporation efficiency = 1300 colonies/261ng DNA = 1300/0.261µg DNA = 4980 colonies/ µg DNA
Machine intermediate 2 = 1400 transformants
Machine intermediate 2 electroporation efficiency = 1400 colonies/241.5ng DNA = 1400/0.2415µg DNA = 5797 colonies/ µg DNA
pNDW5* = 0 transformants (No DNA control)
The pNDW5* vector had no transformants as expected
Electroporation efficiencies were not calculated for Lactodetect and Lactoreport "
"