Team:Uppsala-SwedenWeek16
From 2010.igem.org
(→Characterization of G2) |
(→Characterization of G2) |
||
| (One intermediate revision not shown) | |||
| Line 356: | Line 356: | ||
G2 are the sender cells. H1+Z1 and H2+Z2 are cut-up K1 and K2 constructs without the lower threshold for band detection. In this case, the H1+Z1 and H2+Z2 will show fluorescence only if there is too much AHL. Without AHL, GFP and RFP will be produced constitutively by H1+Z1 and H2+Z2. In the G2 sample, there is no fluorescent plasmid. In the H1+Z1 and H2+Z2 without G2, there should be no AHL in them. Hence, they express GFP and RFP and emit fluorescence. In the G2 samples with H1+Z1 or H2+Z2, there will be excessive amount of G2. So the expected results should be: | G2 are the sender cells. H1+Z1 and H2+Z2 are cut-up K1 and K2 constructs without the lower threshold for band detection. In this case, the H1+Z1 and H2+Z2 will show fluorescence only if there is too much AHL. Without AHL, GFP and RFP will be produced constitutively by H1+Z1 and H2+Z2. In the G2 sample, there is no fluorescent plasmid. In the H1+Z1 and H2+Z2 without G2, there should be no AHL in them. Hence, they express GFP and RFP and emit fluorescence. In the G2 samples with H1+Z1 or H2+Z2, there will be excessive amount of G2. So the expected results should be: | ||
| + | G2: no fluorescence | ||
| + | H1+Z1: green fluorescence | ||
| + | |||
| + | H2+Z2: red fluorescence | ||
| + | |||
| + | G2, H1+Z1: little to no fluorescence | ||
| + | |||
| + | G2, H2+Z2: little to no fluorescence | ||
| + | |||
| + | |||
| + | Which match exactly the obtained results, as shown in the picture below. | ||
[[Image:DSC04430.jpg|600px|thumb|left|From left: G2, H1+Z1, H2+Z2, G2 with H1+Z1, G2 with H2+Z2]] | [[Image:DSC04430.jpg|600px|thumb|left|From left: G2, H1+Z1, H2+Z2, G2 with H1+Z1, G2 with H2+Z2]] | ||
Latest revision as of 00:25, 27 October 2010



Week-16
Start to apply high fidelity PCR due the construct are getting larger and the regular Taq polymerase may not perform as well as the Phusion Polymerase at this length. Based on the company´s manual, we calculated the
|
|
Temperatur |
Time |
Cycle |
|
Initial denaturation |
98 |
30 s |
1 |
|
Denaturation |
98 |
10 |
30 |
|
Annealing |
55.3 |
10s |
30 |
|
Extension |
72 |
1 min 45 s |
30 |
|
Final extension |
72 |
5 min |
1 |
|
|
4 |
forever |
|
The purpose of this was to get more reliable band when doing a PCR and running the gel.
|
Mastermix |
Volume (µL) |
|
H20 |
10.8 |
|
5X phusion HF Buffer |
4 |
|
dNTPs |
2 |
|
VF2 |
1 |
|
VR |
1 |
|
DNA-template |
1 |
|
Phusion Polymerase |
0.2 |
J1C3, J2C3 and J3C1 was picked to build the K4,K5,K6 based on the gelpictures from 27/8/2010. Therefore we do not proceed with the ligated J-construct from the 16/9 due we have good enough samples from August month
Concentration measurement and swithch the backbone from pSB1C3 to psB1A3
|
|
DNA-conc.
|
DNA-vol Digestion |
Restr.Enzym E & P |
FD Buffer 10x |
H20 |
Total |
|
J1C3 |
97.2 |
10.3 |
0.5 & 0.5 |
2 |
6.7 |
20 |
|
J2C3 |
73.6 |
13.6 |
0.5 & 0.5 |
2 |
3.4 |
20 |
|
J3C1 |
71.2 |
14.0 |
0.5 & 0.5 |
2 |
3 |
20 |
|
ccdbA3 |
180.0 |
5 |
0.5 & 0.5 |
2 |
12 |
20 |
Ligation and transformation according the protocol
Phusion colony-PCR for K1, K2, K3, G1, G2 with Phusion Polymerase
We had unsatisfied results with our colonies fluorescence measurement and we suspected that the LacI protein didn’t suppress the fluorescent protein enough. One possible explanation would that BB_P0412 contains BB_C0012 which is the coding region for the LacI protein were degraded too effectively by the LVA degradation tail. As a consequence the R0010, a LacI promoter suppressing the three plasmids flourescent expression, didn’t bind enough LacI and therefore you could detect some flourescence.
Since J-construct contains the BB_R0010 as well, the K4, K5 and K6 construction were postponed.
Therefore we switch the BB_C0012 to another LacI-coding sequence that excluded the degradation tail., called LacIB115 which is in psB1AK3. With the restriction enzymes ApaI and PstI , both restriction sites surround the LacI coding region for psB1AK3 and I-construct. Afterdigestion of both plamids, you run the gel and cut out the band with LacI (without the degradation tail and the I-construction backbone and assembly those two parts.
Characterization of G2
G1 has been characterized by via length confirmation. However, G2 had yet to be confirmed. A simple reporter system of F2621 and I13504 (GFP gene) was constructed. However, the reporter system failed to be transformed into cells. In order to confirm G2 some other way, two few improvisational constructs were made, codenamed Z1 and Z2:
Z1: P0412-R0010-I13504
Z2: P0412-R0010-I13507
Z1 and Z2 produce GFP and RFP constitutively. If coupled with H1 and H2 respectively, they obtain an upper threshold for their band-detection, but not the lower threshold.
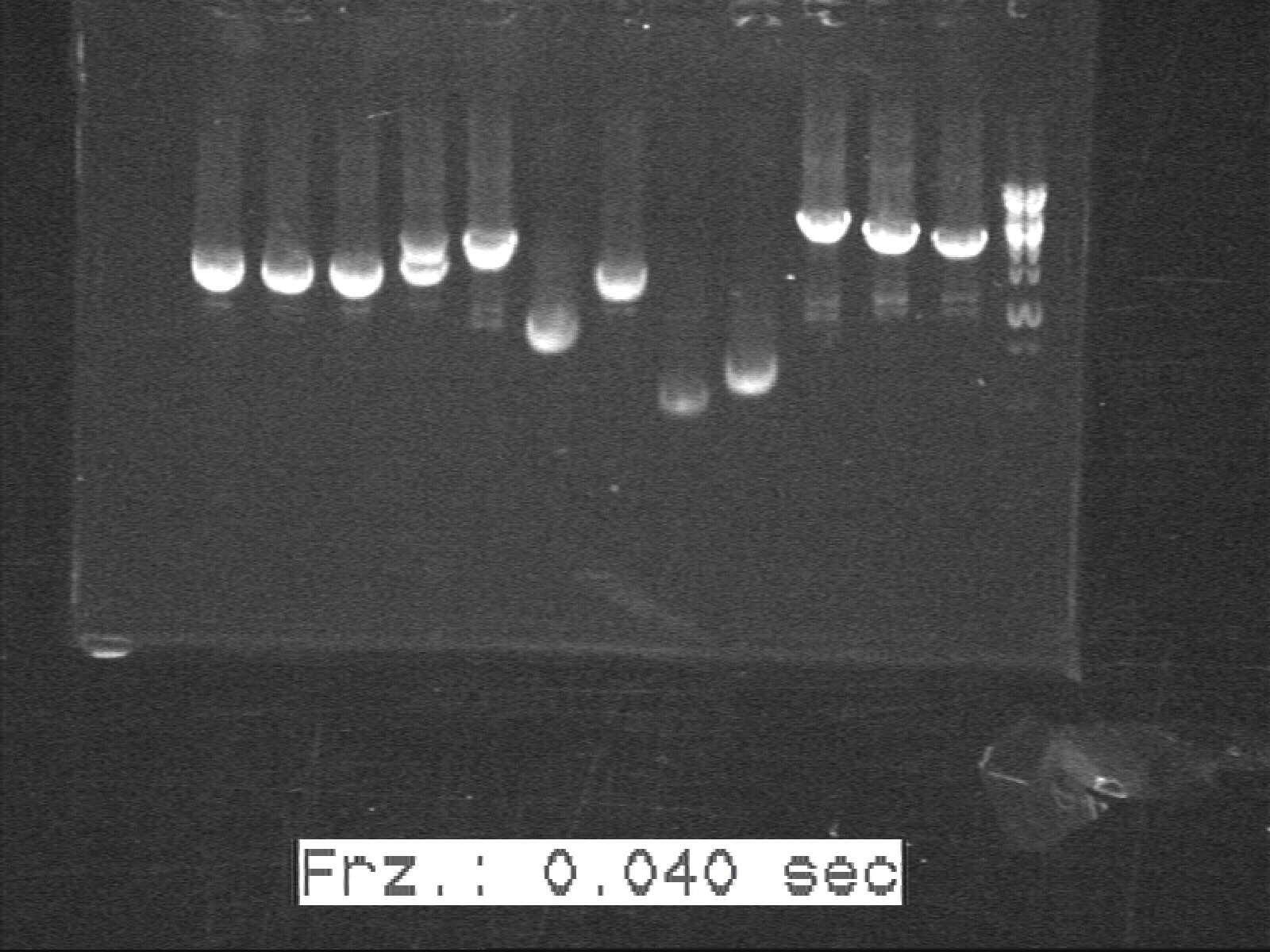
5 LB cultures were prepared to test our G2. They are: G2, H1+Z1, H2+Z2, G2 with H1+Z1, and G2 with H2+Z2. All the LB cultures were inoculated at the same time, in 6 ml LB in culture tubes. The cultures grew overnight in 37°C under 250 rpm.
G2 are the sender cells. H1+Z1 and H2+Z2 are cut-up K1 and K2 constructs without the lower threshold for band detection. In this case, the H1+Z1 and H2+Z2 will show fluorescence only if there is too much AHL. Without AHL, GFP and RFP will be produced constitutively by H1+Z1 and H2+Z2. In the G2 sample, there is no fluorescent plasmid. In the H1+Z1 and H2+Z2 without G2, there should be no AHL in them. Hence, they express GFP and RFP and emit fluorescence. In the G2 samples with H1+Z1 or H2+Z2, there will be excessive amount of G2. So the expected results should be:
G2: no fluorescence
H1+Z1: green fluorescence
H2+Z2: red fluorescence
G2, H1+Z1: little to no fluorescence
G2, H2+Z2: little to no fluorescence
Which match exactly the obtained results, as shown in the picture below.
 "
"