Team:Uppsala-SwedenWeek11
From 2010.igem.org
(New page: {{Template:Uppsala}}<!--Do not remove the first and last lines in this page!--> == Week-11 ==) |
(→Week-11) |
||
| Line 1: | Line 1: | ||
{{Template:Uppsala}}<!--Do not remove the first and last lines in this page!--> | {{Template:Uppsala}}<!--Do not remove the first and last lines in this page!--> | ||
== Week-11 == | == Week-11 == | ||
| + | |||
| + | == Construction of H1- G8 == | ||
| + | |||
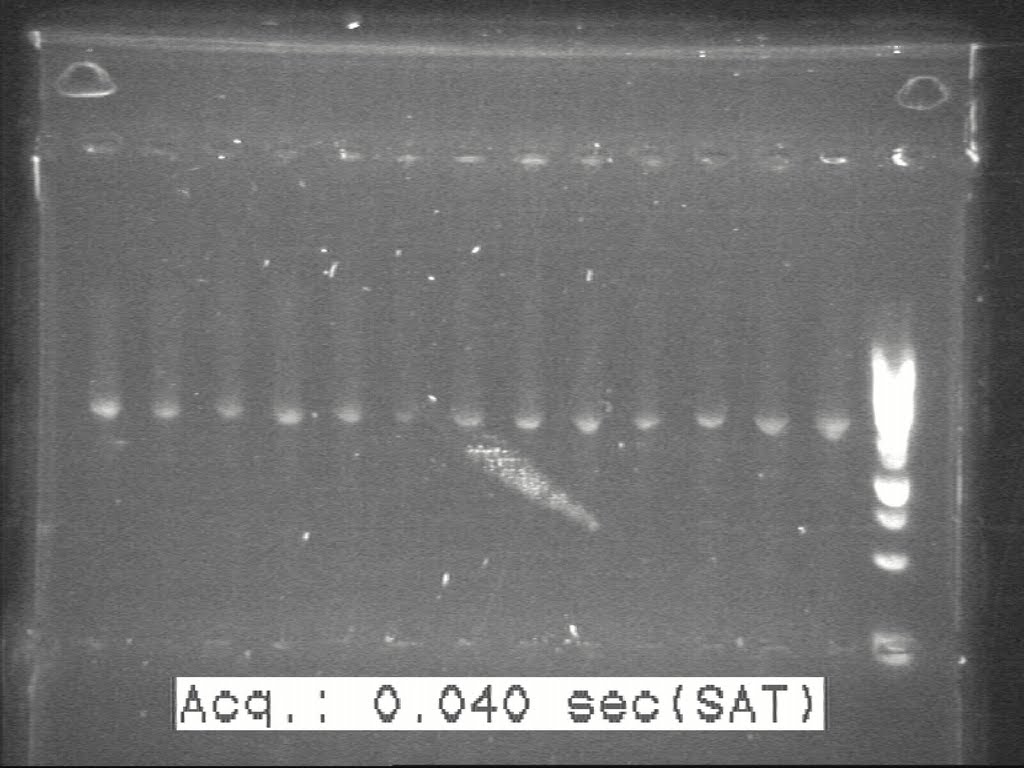
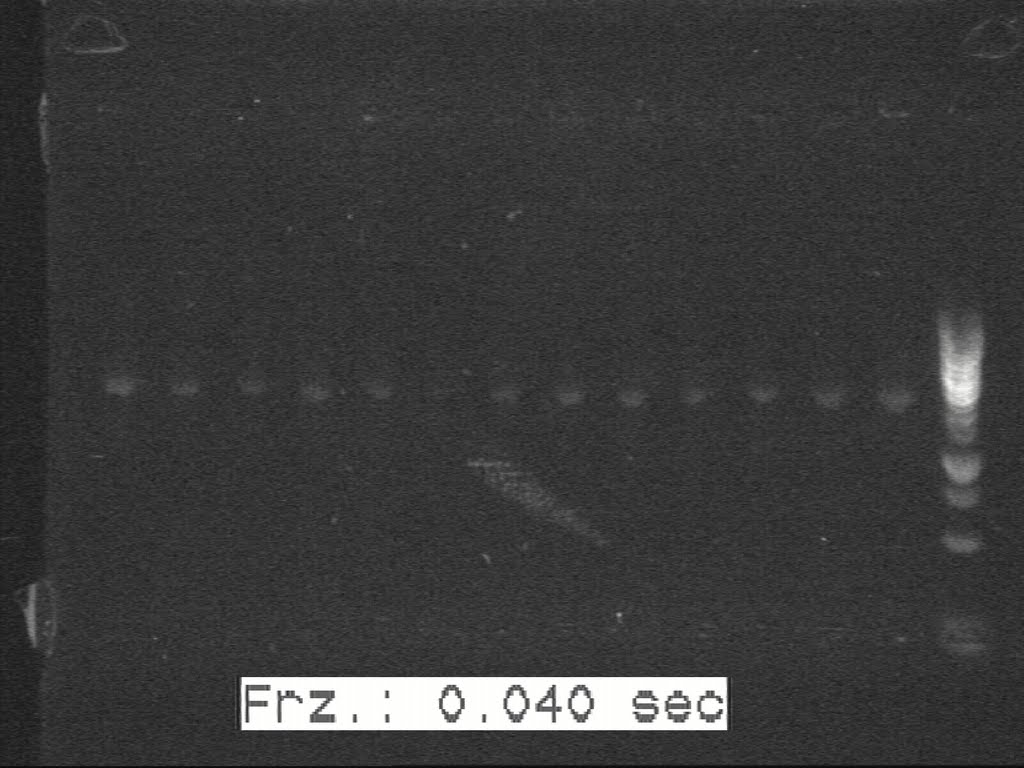
| + | 5 colonies were picked from each of those plates and sent to perform c-PCR. According to the band size on gel picture, 1 or 2 colonies of each set were selected to inoculate. | ||
| + | |||
| + | [[Image:20100817 H1-H2-H8 c-PCR 20100814.jpg|500px|thumb|center|border|H1-H2-H8]] | ||
| + | |||
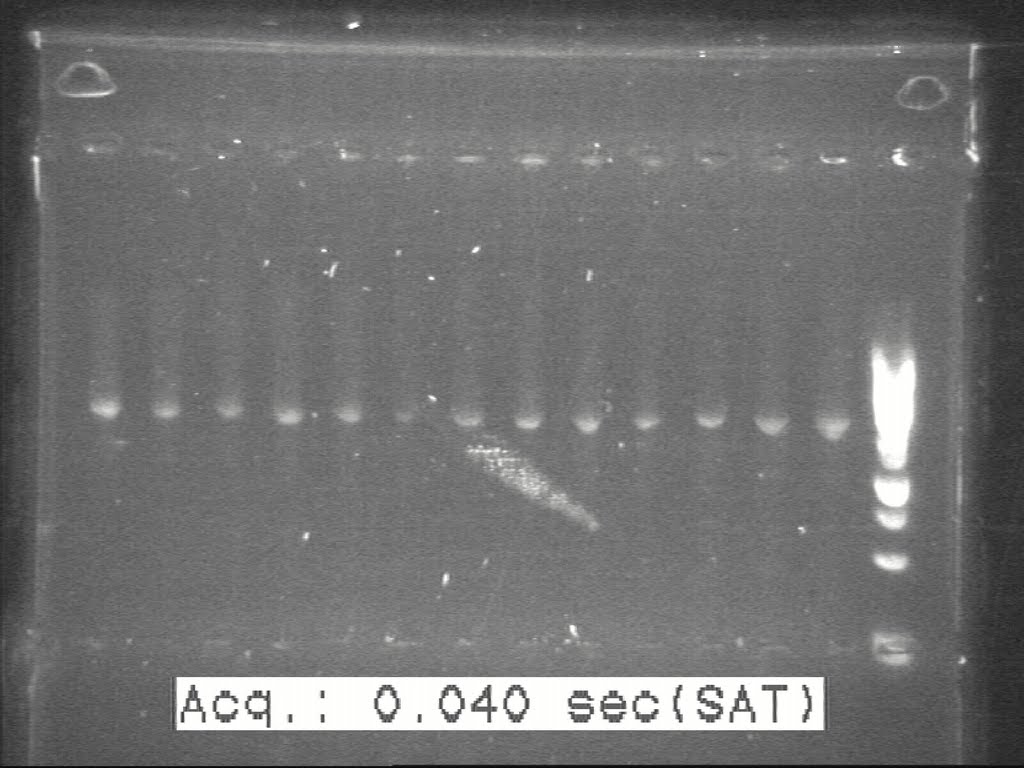
| + | From the above gel picture it was not very clear. Some fiddling around with the controls and we got a much better image (given below). | ||
| + | |||
| + | [[Image:20100817 H1-H2-H8 c-PCR 20100814(2).jpg|500px|thumb|center|border|H1-H2-H8 with different illumination to identify the constructs]] | ||
| + | |||
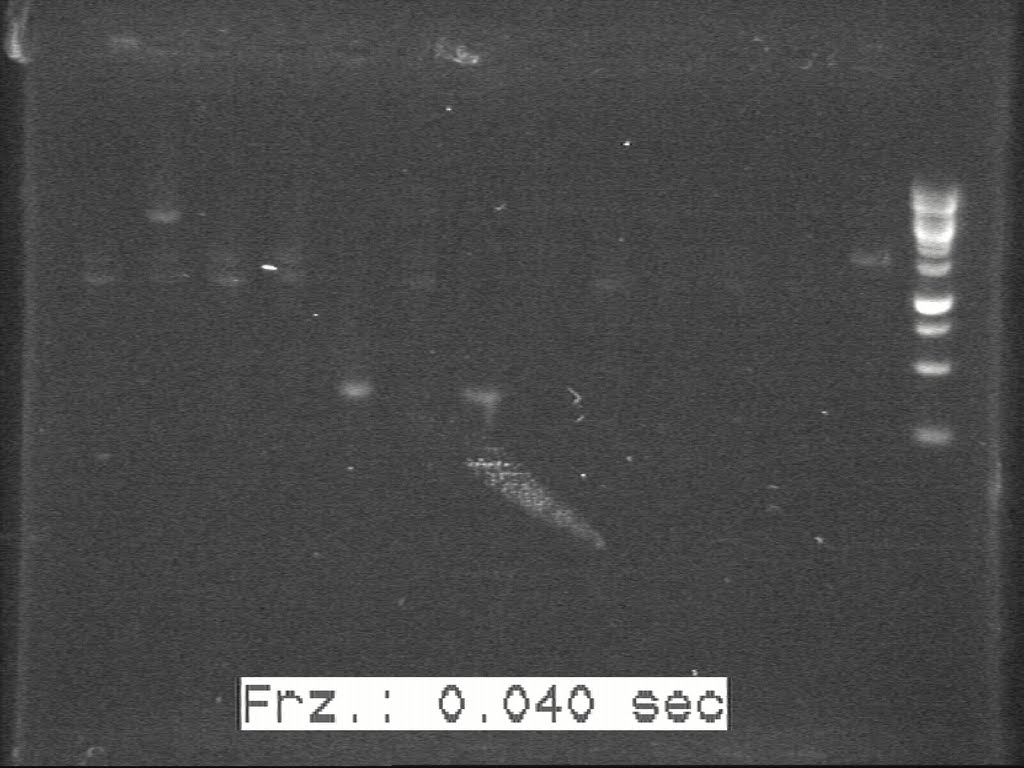
| + | [[Image:20100817 H3--H4 c-PCR 20100814.jpg|500px|thumb|center|border|H3-H4]] | ||
| + | |||
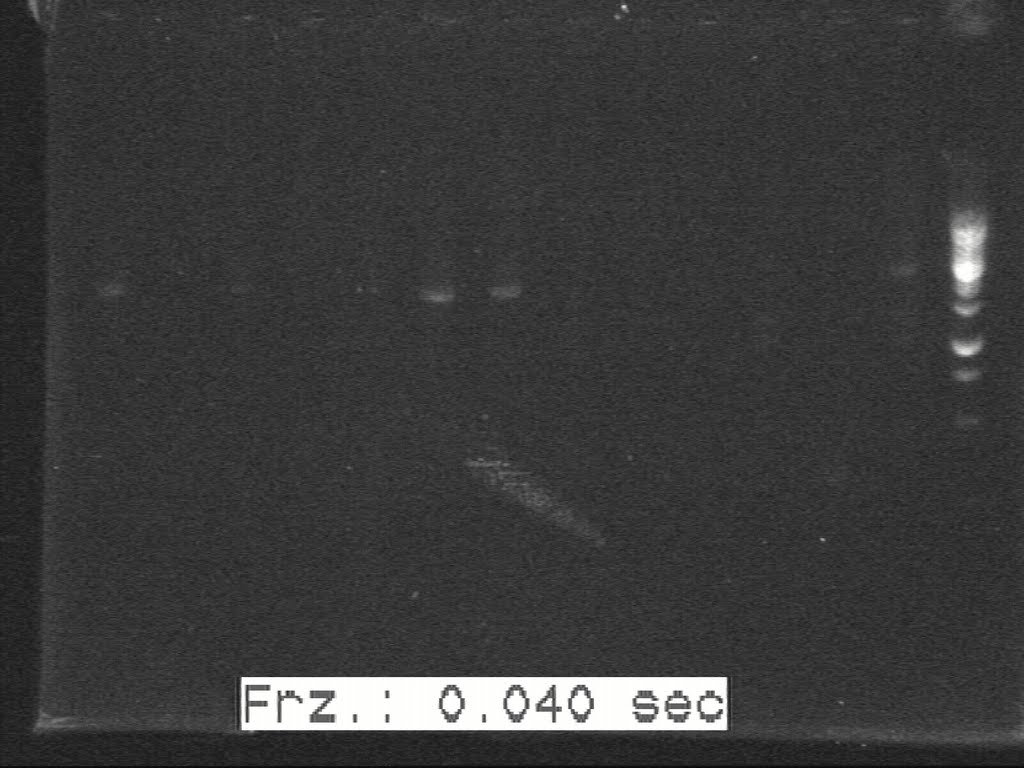
| + | [[Image:20100817 H5-H6 c-PCR 20100814(3).jpg|500px|thumb|center|border|H5-H6]] | ||
| + | |||
| + | [[Image:20100817 H7-H8 c-PCR 20100814(3).jpg|500px|thumb|center|border|H7-H8]] | ||
| + | |||
| + | The samples selected in Colony PCR were inoculated for obtaining material to do plasmid extraction. Overnight culture was used to make glycerol stocks and plasmids were extracted. The plasmid concentration was measured for estimating the amount of enzymes required for digestion and ligation. | ||
| + | |||
| + | Plasmids were digested with proper enzymes bought from Sigma-Aldrich and run for gel to confirm the size. The plasmid parts where also digested for use in ligation for the next step.The correct set of parts was ligated using Ampicillin backbone via 3A assembly following the Flowchart as per the plan. | ||
Latest revision as of 18:10, 26 October 2010



Week-11
Construction of H1- G8
5 colonies were picked from each of those plates and sent to perform c-PCR. According to the band size on gel picture, 1 or 2 colonies of each set were selected to inoculate.
From the above gel picture it was not very clear. Some fiddling around with the controls and we got a much better image (given below).
The samples selected in Colony PCR were inoculated for obtaining material to do plasmid extraction. Overnight culture was used to make glycerol stocks and plasmids were extracted. The plasmid concentration was measured for estimating the amount of enzymes required for digestion and ligation.
Plasmids were digested with proper enzymes bought from Sigma-Aldrich and run for gel to confirm the size. The plasmid parts where also digested for use in ligation for the next step.The correct set of parts was ligated using Ampicillin backbone via 3A assembly following the Flowchart as per the plan.
 "
"